3Q4I
 
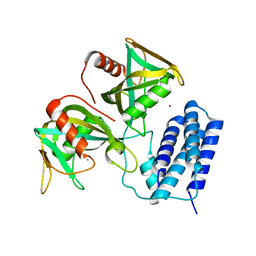 | | Crystal structure of CDP-Chase in complex with Gd3+ | | Descriptor: | GADOLINIUM ION, Phosphohydrolase (MutT/nudix family protein) | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
5U17
 
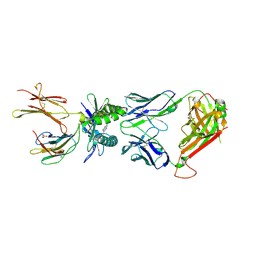 | |
5U2V
 
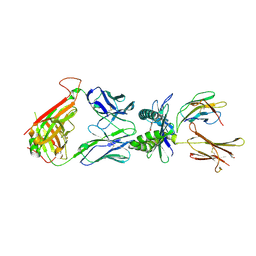 | |
5WI4
 
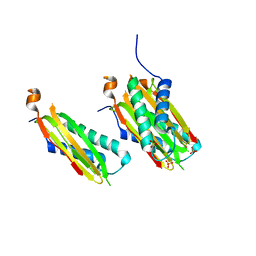 | | CRYSTAL STRUCTURE OF DYNLT1/TCTEX-1 IN COMPLEX WITH ARHGEF2 | | Descriptor: | Dynein light chain Tctex-type 1,Rho guanine nucleotide exchange factor 2, SULFATE ION | | Authors: | Balan, M, Ishiyama, N, Marshall, C.B, Ikura, M. | | Deposit date: | 2017-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity.
Sci Signal, 10, 2017
|
|
3TPE
 
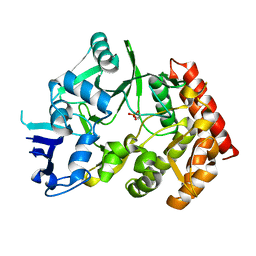 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPD
 
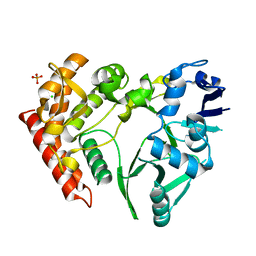 | | Structure of pHipA, monoclinic form | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMR
 
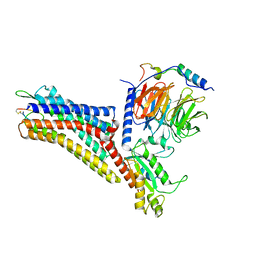 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7K8L
 
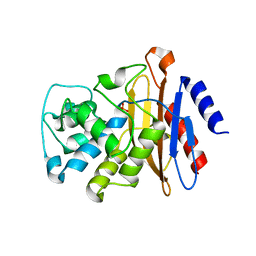 | | Beta-lactamase, Unmixed | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8000102 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8E
 
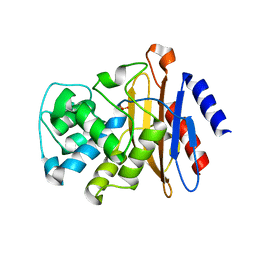 | | Beta-lactamase mixed with Ceftriaxone, 5ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40005636 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8H
 
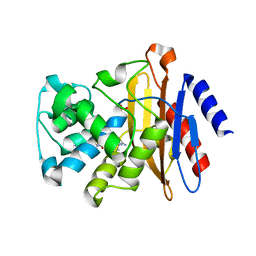 | | Beta-lactamase mixed with Ceftriaxone, 50ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60006261 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8K
 
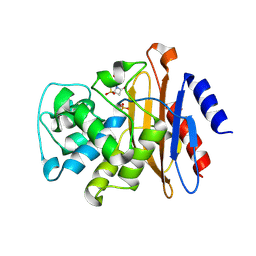 | | Beta-lactamase mixed with Sulbactam, 60ms | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULBACTAM, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8F
 
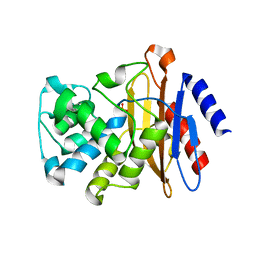 | | Beta-lactamase mixed with Ceftriaxone, 10ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60003138 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
5ZF2
 
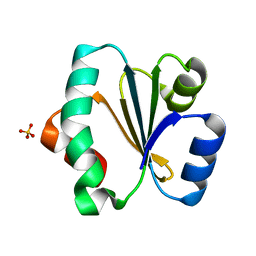 | | Crystal structure of Trxlp from Edwardsiella tarda EIB202 | | Descriptor: | SULFATE ION, Thioredoxin (H-type,TRX-H) | | Authors: | Yang, C, Quan, S. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Edwardsiella piscicida thioredoxin-like protein inhibits ASK1-MAPKs signaling cascades to promote pathogenesis during infection.
Plos Pathog., 15, 2019
|
|
6A0A
 
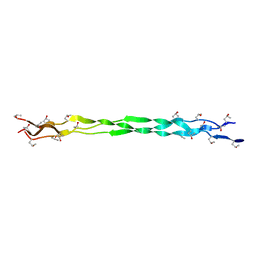 | |
6A0C
 
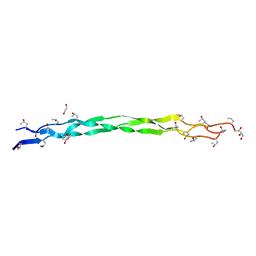 | | Structure of a triple-helix region of human collagen type III | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, collagen type III peptide | | Authors: | Yang, X, Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization by high-resolution crystal structure analysis of a triple-helix region of human collagen type III with potent cell adhesion activity.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
8JVD
 
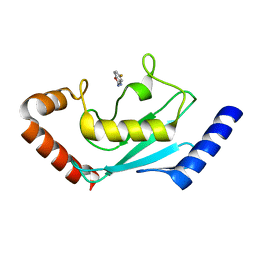 | |
8JUC
 
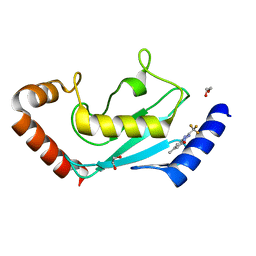 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
8JRR
 
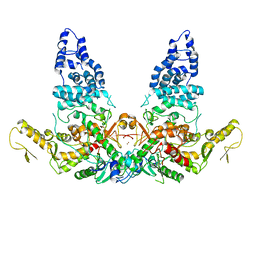 | | Structure of E6AP-E6 complex in Det2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRQ
 
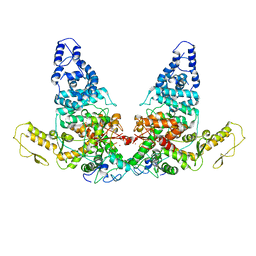 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRN
 
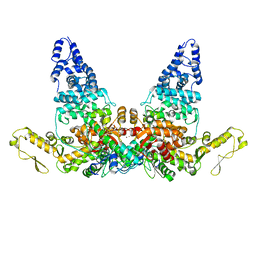 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRP
 
 | | Structure of E6AP-E6 complex in Att3 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
