3VPJ
 
 | |
7Y20
 
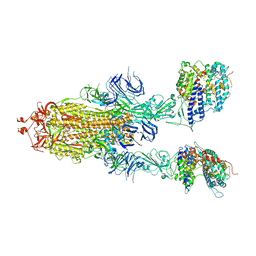 | | S-ECD (Omicron BA.3) in complex with two PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7Y21
 
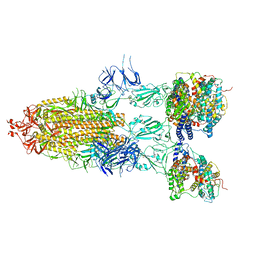 | | S-ECD (Omicron BA.5) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
3UCX
 
 | |
3V3L
 
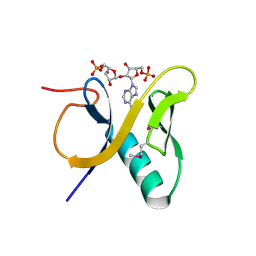 | | Crystal structure of human RNF146 WWE domain in complex with iso-ADPRibose | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146 | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2011-12-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination.
Genes Dev., 26, 2012
|
|
3W8I
 
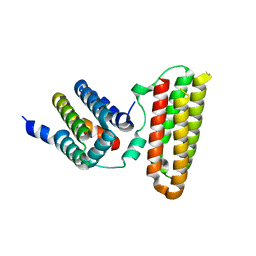 | |
3TL3
 
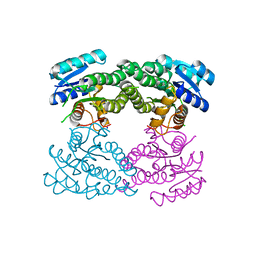 | |
3TRR
 
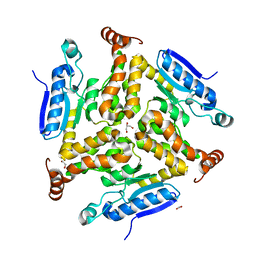 | |
3W8H
 
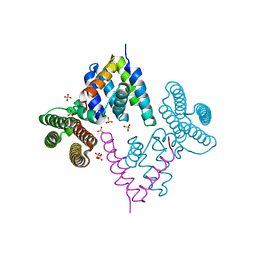 | |
3WA5
 
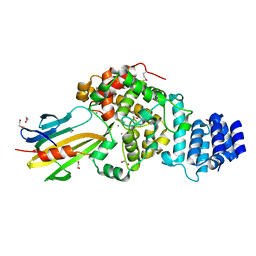 | | Crystal Structure of type VI peptidoglycan muramidase effector Tse3 in complex with its cognate immunity protein Tsi3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Tse3-specific immunity protein, ... | | Authors: | Ding, J, Wang, T, Liu, W, Wang, D.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex structure of type VI peptidoglycan muramidase effector and a cognate immunity protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5Y1H
 
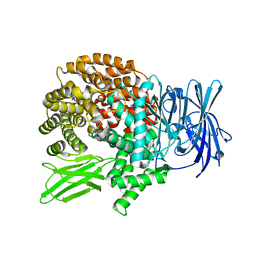 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[[2,4-bis(fluoranyl)phenyl]methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y3I
 
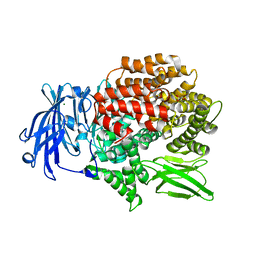 | |
5Y1S
 
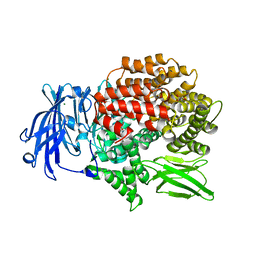 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, (2S)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1V
 
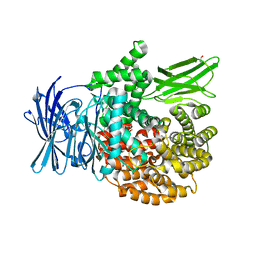 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(2,6-diethylphenyl)carbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide
To Be Published
|
|
7CGE
 
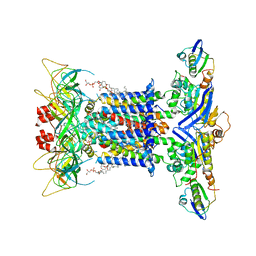 | | The overall structure of nucleotide free MlaFEDB complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CGN
 
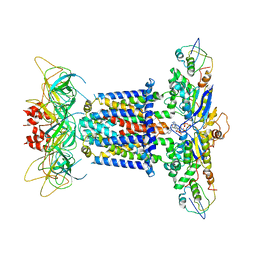 | | The overall structure of the MlaFEDB complex in ATP-bound EQtall conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
5Y19
 
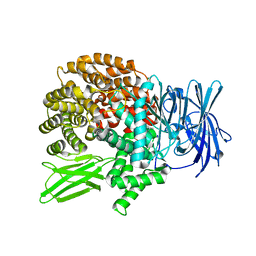 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)- N-hydroxy-4-methylpentanamide. | | Descriptor: | (2S)-2-[[(2S,3R)-3-azanyl-2-oxidanyl-4-phenyl-butanoyl]amino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)- N-hydroxy-4-methylpentanamide.
To Be Published
|
|
7CH0
 
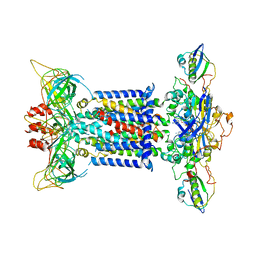 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
4B9D
 
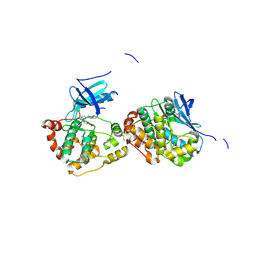 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4APC
 
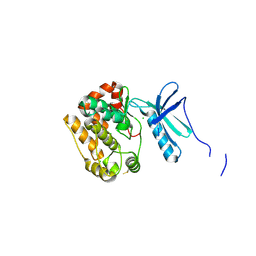 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
2XWC
 
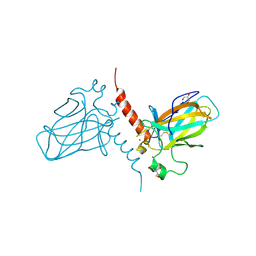 | | Crystal structure of the DNA binding domain of human TP73 refined at 1.8 A resolution | | Descriptor: | GLYCEROL, TRIS(HYDROXYETHYL)AMINOMETHANE, TUMOUR PROTEIN P73, ... | | Authors: | Canning, P, Zhang, Y, Vollmar, M, Krojer, T, Ugochukwu, E, Muniz, J.R.C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
8IBU
 
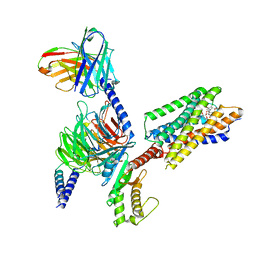 | | Cryo-EM structure of the erythromycin-bound motilin receptor-Gq protein complex | | Descriptor: | ERYTHROMYCIN A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | You, C, Jiang, Y, Xu, H.E, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
8IBV
 
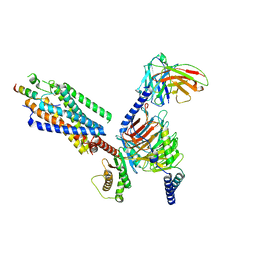 | | Cryo-EM structure of the motilin-bound motilin receptor-Gq protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Jiang, Y, Xu, H.E, You, C, Xu, Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
7WZW
 
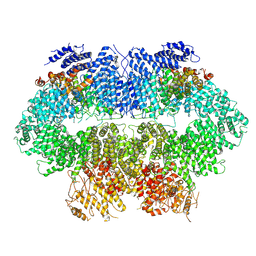 | | Cryo-EM structure of MEC1-DDC2-MMS | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
7WZR
 
 | | Cryo-EM structure of Mec1-HU | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
