7ZQI
 
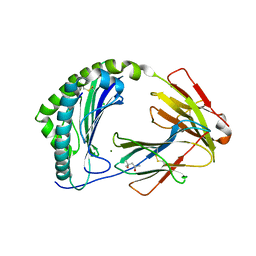 | | MHC class I from a wild bird in complex with a nonameric peptide P2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
7ZQJ
 
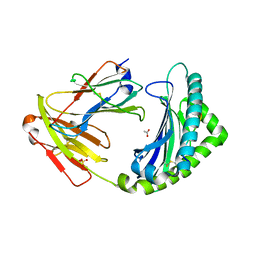 | | MHC class I from a wild bird in complex with a nonameric peptide P3 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Eltschkner, S, Mellinger, S, Buus, S, Nielsen, M, Paulsson, K.M, Lindkvist-Petersson, K, Westerdahl, H. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of songbird MHC class I reveals antigen binding that is flexible at the N-terminus and static at the C-terminus.
Front Immunol, 14, 2023
|
|
8CHA
 
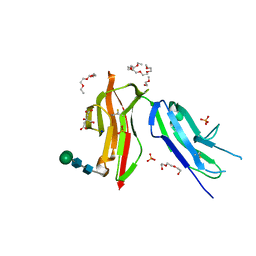 | | Fc gamma RIIa 27W/131H variant ectodomain | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Foy, E.G, Thomsen, M, Goldman, A, Robinson, J.I. | | Deposit date: | 2023-02-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fc gamma RIIa 27W/131H variant ectodomain
To Be Published
|
|
8CI5
 
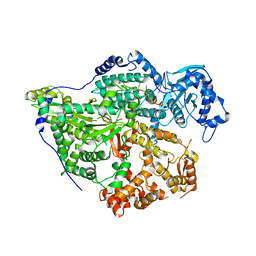 | | Structure of the SNV L protein bound to 5' RNA | | Descriptor: | RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Meier, K, Thorkelsson, S.R, Durieux Trouilleton, Q, Vogel, D, Yu, D, Kosinski, J, Cusack, S, Malet, H, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of the Sin Nombre virus L protein.
Plos Pathog., 19, 2023
|
|
4J7X
 
 | |
6H2B
 
 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
6TRT
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant S180C/T742C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TERBIUM(III) ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
4NPX
 
 | | Structure of hypothetical protein Cj0539 from Campylobacter jejuni | | Descriptor: | Putative uncharacterized protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Adkins, J.N, Endres, M, Nissen, M, Konkel, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-11-22 | | Release date: | 2014-01-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of hypothetical protein Cj0539 from Campylobacter jejuni
To be Published
|
|
6TS8
 
 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) double cysteine mutant G177C/A786C. | | Descriptor: | UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Ibba, R, Hensen, M, Chandran, A. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6QHG
 
 | | Structure of the cap-binding domain of Rift Valley Fever virus L protein | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Gogrefe, N, Reindl, S, Gunther, S, Rosenthal, M. | | Deposit date: | 2019-01-16 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-05 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | Structure of a functional cap-binding domain in Rift Valley fever virus L protein.
Plos Pathog., 15, 2019
|
|
1CPY
 
 | | SITE-DIRECTED MUTAGENESIS ON (SERINE) CARBOXYPEPTIDASE Y FROM YEAST. THE SIGNIFICANCE OF THR 60 AND MET 398 IN HYDROLYSIS AND AMINOLYSIS REACTIONS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE | | Authors: | Sorensen, S.B, Raaschou-Nielsen, M, Mortensen, U, Remington, S.J, Breddam, K. | | Deposit date: | 1995-03-24 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Site-Directed Mutagenesis on (Serine) Carboxypeptidase Y from Yeast. The Significance of Thr 60 and met 398 in Hydrolysis and Aminolysis Reactions
J.Am.Chem.Soc., 117, 1995
|
|
3QHS
 
 | | Crystal structure of full-length Hfq from Escherichia coli | | Descriptor: | Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Sjoeblom, B, Blaesi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of full-length Hfq from Escherichia coli
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3QO3
 
 | | Crystal structure of Escherichia coli Hfq, in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein hfq | | Authors: | Beich-Frandsen, M, Vecerek, B, Hammele, H, Kloiber, K, Sjoeblom, B, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical studies on ATP binding and hydrolysis by the Escherichia coli RNA chaperone Hfq
Plos One, 7, 2012
|
|
1BL1
 
 | | PTH RECEPTOR N-TERMINUS FRAGMENT, NMR, 1 STRUCTURE | | Descriptor: | PARATHYROID HORMONE RECEPTOR | | Authors: | Pellegrini, M, Bisello, A, Rosenblatt, M, Chorev, M, Mierke, D.F. | | Deposit date: | 1998-07-22 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding domain of human parathyroid hormone receptor: from conformation to function.
Biochemistry, 37, 1998
|
|
3ST3
 
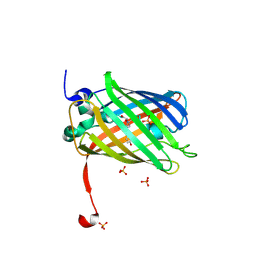 | | Dreiklang - off state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3BFT
 
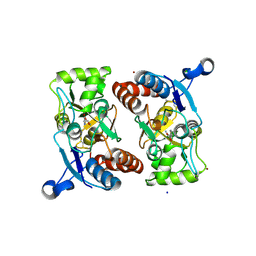 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3BFU
 
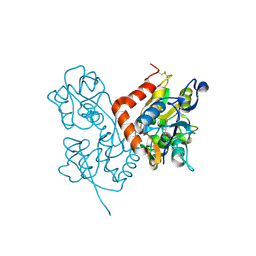 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
4DVY
 
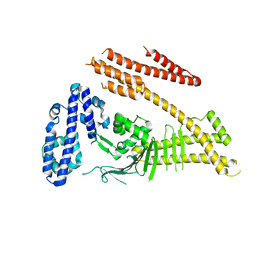 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
1B6Y
 
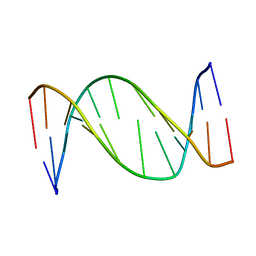 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
3Q2S
 
 | | Crystal Structure of CFIm68 RRM/CFIm25 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Yang, Q, Coseno, M, Gilmartin, G.M, Doublie, S. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Structure, 19, 2011
|
|
3Q2T
 
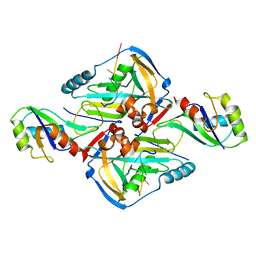 | | Crystal Structure of CFIm68 RRM/CFIm25/RNA complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6, RNA | | Authors: | Yang, Q, Coseno, M, Gilmartin, G.M, Doublie, S. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.061 Å) | | Cite: | Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Structure, 19, 2011
|
|
4N1L
 
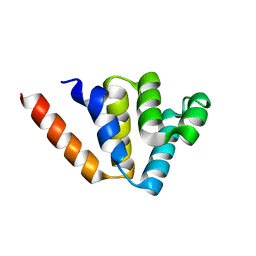 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1K
 
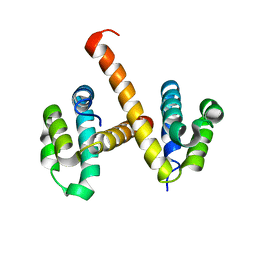 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N1J
 
 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1B5K
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
