2IZC
 
 | | APOSTREPTAVIDIN PH 2.0 I222 COMPLEX | | Descriptor: | CHLORIDE ION, SODIUM ION, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZI
 
 | | STREPTAVIDIN-BIOTIN PH 2.53 I4122 STRUCTURE | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZH
 
 | | STREPTAVIDIN-BIOTIN PH 10.44 I222 COMPLEX | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
9M0E
 
 | | Enhancing the synthesis efficiency of galacto-oligosaccharides of a beta-galactosidase from Paenibacillus barengoltzii by engineering the active and distal sites | | Descriptor: | Beta-galactosidase | | Authors: | Yu, H.Y, Wang, Y.L, Yang, Z.S, Liu, X, Xin, F.J. | | Deposit date: | 2025-02-24 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Enhancing the synthesis efficiency of galacto-oligosaccharides of a beta-galactosidase from Paenibacillus barengoltzii by engineering the active and distal sites.
Food Chem, 483, 2025
|
|
7N5U
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 21) | | Descriptor: | DNA Strain II, DNA Strand I, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5W
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
9O63
 
 | | Crystal structure of PLK4 and RP1664 complex | | Descriptor: | 5-cyclopropyl-N~2~-[2,6-difluoro-4-(methanesulfonyl)phenyl]-N~2~-methyl-6-(1-methyl-1H-imidazol-4-yl)-N~4~-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Maderova, Z, Zimmermann, M, Sicheri, F. | | Deposit date: | 2025-04-11 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of RP-1664: A First-in-Class Orally Bioavailable, Selective PLK4 Inhibitor.
J.Med.Chem., 68, 2025
|
|
6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
8QQI
 
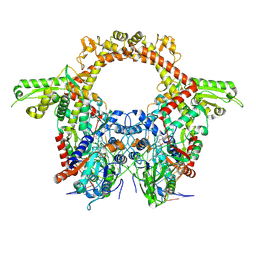 | | E.coli DNA gyrase in complex with 217 bp substrate DNA and LEI-800 | | Descriptor: | DNA gyrase subunit A, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Ghilarov, D, Martin, N.I, van der Stelt, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of isoquinoline sulfonamides as allosteric gyrase inhibitors with activity against fluoroquinolone-resistant bacteria.
Nat.Chem., 16, 2024
|
|
4YNE
 
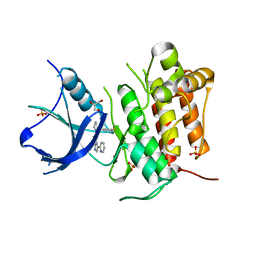 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YMJ
 
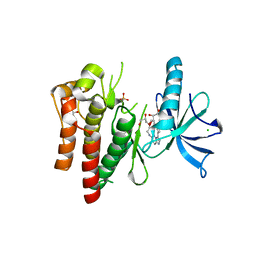 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YPS
 
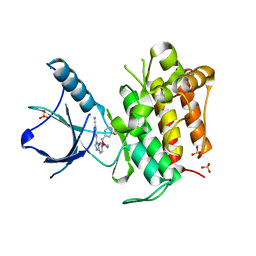 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-{6-[(3R)-3-(3-fluorophenyl)morpholin-4-yl]imidazo[1,2-b]pyridazin-3-yl}benzonitrile, High affinity nerve growth factor receptor, SULFATE ION | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1012 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
2H96
 
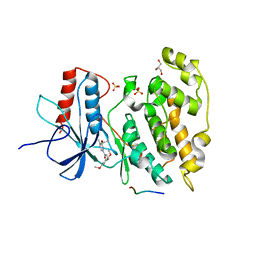 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
4Z9P
 
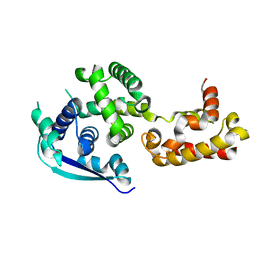 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
5KX5
 
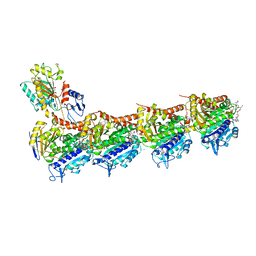 | | Crystal structure of tubulin-stathmin-TTL-Compound 11 complex | | Descriptor: | (2~{S},4~{R})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-3-[[(2~{S},3~{S})-2-[[(2~{R})-1,2-dimethylpyrrolidin-2-yl]carbonylamino]-3-methyl-pentanoyl]-methyl-amino]-4-methyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-(4-aminophenyl)-2-methyl-pentanoic acid, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Parris, K. | | Deposit date: | 2016-07-20 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis, and Cytotoxic Evaluation of Novel Tubulysin Analogues as ADC Payloads.
ACS Med Chem Lett, 7, 2016
|
|
2GMX
 
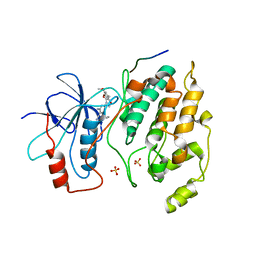 | | Selective Aminopyridine-Based C-Jun N-terminal Kinase inhibitors with cellular activity | | Descriptor: | C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, N-(4-AMINO-5-CYANO-6-ETHOXYPYRIDIN-2-YL)-2-(4-BROMO-2,5-DIMETHOXYPHENYL)ACETAMIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-04-07 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
J.Med.Chem., 49, 2006
|
|
6LNZ
 
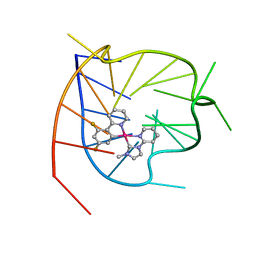 | |
8R06
 
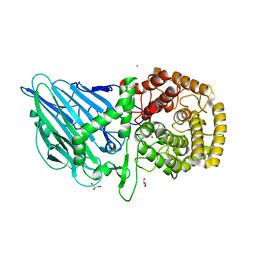 | | CRYSTAL STRUCTURE OF THERMOANAEROBACTERIUM XYLANOLYTICUM GH116 BETA-GLUCOSIDASE WITH A COVALENTLY BOUND CYCLOPHELLITOL AZIRIDINE | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{R},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Davies, G.J, Breen, I.Z. | | Deposit date: | 2023-10-30 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOANAEROBACTERIUM XYLOLYTICUM GH116 BETA-GLUCOSIDASE WITH A COVALENTLY BOUND CYCLOPHELLITOL AZIRIDINE
JACS, 40, 2017
|
|
8D6E
 
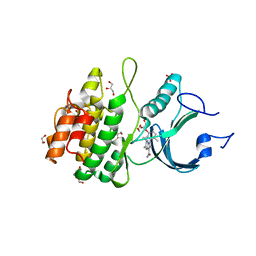 | | Crystal Structure of Human Myt1 Kinase domain Bounded with RP-6306 | | Descriptor: | (1P)-2-amino-1-(3-hydroxy-2,6-dimethylphenyl)-5,6-dimethyl-1H-pyrrolo[2,3-b]pyridine-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6D
 
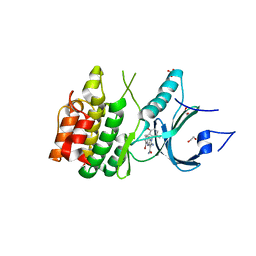 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 39 | | Descriptor: | (1P)-2-amino-5-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6F
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with Eph receptor inhibitor / compound 41 | | Descriptor: | (1M)-2-amino-1-(5-hydroxy-2-methylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, SULFATE ION | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6C
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 28 | | Descriptor: | (1P)-2-amino-6-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
1CN4
 
 | |
9NBB
 
 | | Augmin/V junction(closed) | | Descriptor: | AUGMIN subunit 2, AUGMIN subunit 3, AUGMIN subunit 5, ... | | Authors: | Ashaduzzaman, M, Al-Bassam, J, Taheri, A. | | Deposit date: | 2025-02-13 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.
Biorxiv, 2025
|
|
