5WYD
 
 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
8Y86
 
 | | Human AE3 with NaHCO3- | | Descriptor: | Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8YFU
 
 | | Cryo EM structure of human phosphate channel XPR1 at intermediate state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YEX
 
 | | Cryo EM structure of human phosphate channel XPR1 at apo state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YF4
 
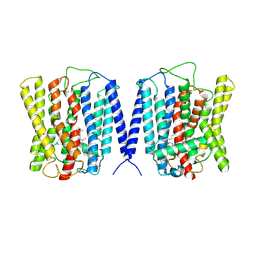 | | Cryo EM structure of human phosphate channel XPR1 at open and inward-facing state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-24 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YFD
 
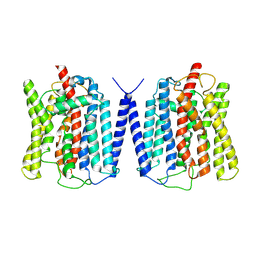 | | Cryo EM structure of human phosphate channel XPR1 at open state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-24 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YET
 
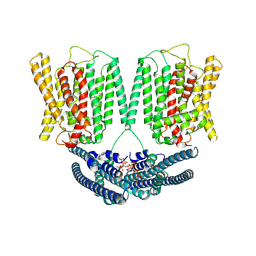 | | Cryo EM structure of human phosphate channel XPR1 in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YFX
 
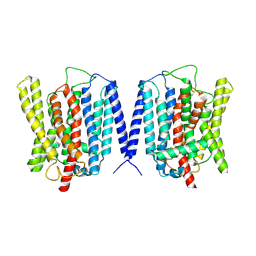 | | Cryo EM structure of human phosphate channel XPR1 at inward-facing state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
8YFW
 
 | | Cryo EM structure of human phosphate channel XPR1 at intermediate state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
9IWS
 
 | | Cryo EM structure of human phosphate channel XPR1 in complex with IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-07-25 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 2024
|
|
6LLG
 
 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | Descriptor: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | Authors: | Wang, X, Liu, M. | | Deposit date: | 2019-12-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
6LLZ
 
 | |
6LLW
 
 | |
6R5W
 
 | | Crystal structure of the receptor binding protein (gp15) of Listeria phage PSA | | Descriptor: | ACETATE ION, CADMIUM ION, Gp15 protein, ... | | Authors: | Dunne, M, Ernst, P, Pluckthun, A, Loessner, M.J, Kilcher, S. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins.
Cell Rep, 29, 2019
|
|
7XDX
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDV
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDW
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDY
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
4XI3
 
 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with Bazedoxifene | | Descriptor: | Bazedoxifene, Estrogen receptor | | Authors: | Fanning, S.W, Mayne, C.G, Toy, W, Carlson, K, Greene, B, Nowak, J, Walter, R, Panchamukhi, S, Tajhorshid, E, Nettles, K.W, Chandarlapaty, S, Katzenellenbogen, J, Greene, G.L. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | The SERM/SERD bazedoxifene disrupts ESR1 helix 12 to overcome acquired hormone resistance in breast cancer cells.
Elife, 7, 2018
|
|
8SYC
 
 | | Crystal structure of PDE3B in complex with GSK4394835A | | Descriptor: | MAGNESIUM ION, [3-[(4,7-dimethoxyquinolin-2-yl)carbonylamino]-5-[methyl-(phenylmethyl)carbamoyl]phenyl]-oxidanyl-oxidanylidene-boron, cGMP-inhibited 3',5'-cyclic phosphodiesterase 3B | | Authors: | Concha, N.O, Nolte, R. | | Deposit date: | 2023-05-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and SAR Study of Boronic Acid-Based Selective PDE3B Inhibitors from a Novel DNA-Encoded Library.
J.Med.Chem., 67, 2024
|
|
1VYU
 
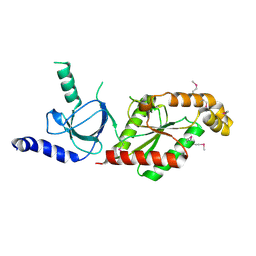 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYV
 
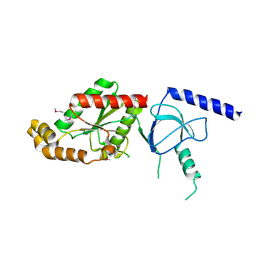 | | beta4 subunit of Ca2+ channel | | Descriptor: | CALCIUM CHANNEL BETA-4SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYT
 
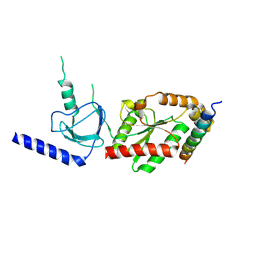 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
7Y1Y
 
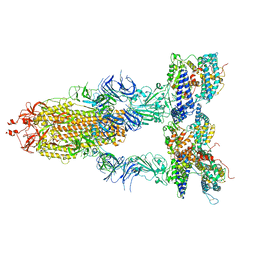 | | S-ECD (Omicron BA.2) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7Y1Z
 
 | | S-ECD (Omicron BA.3) in complex with three PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
