1DZE
 
 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
2D9Q
 
 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5YFI
 
 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-21 | | Release date: | 2018-12-05 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | Descriptor: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-09-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
6JIG
 
 | | Crystal structure of GMP reductase C318A from Trypanosoma brucei in complex with guanosine 5'-monophosphate | | Descriptor: | GMP reductase, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION | | Authors: | Mase, H, Imamura, A, Nishimura, S, Inui, T. | | Deposit date: | 2019-02-21 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Allosteric regulation accompanied by oligomeric state changes of Trypanosoma brucei GMP reductase through cystathionine-beta-synthase domain.
Nat Commun, 11, 2020
|
|
8I34
 
 | | The crystal structure of EPD-BCP1 from a marine sponge | | Descriptor: | (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-4,8,13,17-tetramethyl-3-oxidanyl-19-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]-1-[(1~{R},4~{S})-1,2,2-trimethyl-4-oxidanyl-cyclopentyl]nonadeca-2,4,6,8,10,12,14,16-octaen-18-yn-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASTAXANTHIN, ... | | Authors: | Shomura, Y, Kawasaki, S. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | An ependymin-related blue carotenoprotein decorates marine blue sponge.
J.Biol.Chem., 299, 2023
|
|
6JL8
 
 | |
5YWY
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | Descriptor: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
4YD9
 
 | | Crystal structure of squid hemocyanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CU2-O2 CLUSTER, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matsuno, A, Gai, Z, Kato, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-02-21 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 3.8-MDa Respiratory Supermolecule Hemocyanin at 3.0 angstrom Resolution
Structure, 23, 2015
|
|
6LYC
 
 | | Crystal structure of the NOD SIRPa complex with D4-2 | | Descriptor: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | Authors: | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
7W9Q
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
7W9R
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-18 | | Descriptor: | (2~{R})-2-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
3TXN
 
 | |
3TXM
 
 | |
2RGH
 
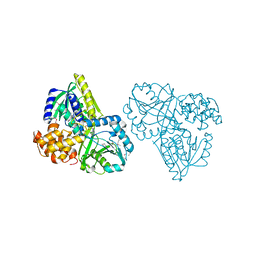 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-01 | | Release date: | 2008-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
2RGO
 
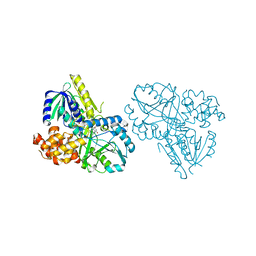 | | Structure of Alpha-Glycerophosphate Oxidase from Streptococcus sp.: A Template for the Mitochondrial Alpha-Glycerophosphate Dehydrogenase | | Descriptor: | Alpha-Glycerophosphate Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Colussi, T, Boles, W, Mallett, T.C, Karplus, P.A, Claiborne, A. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of alpha-glycerophosphate oxidase from Streptococcus sp.: a template for the mitochondrial alpha-glycerophosphate dehydrogenase.
Biochemistry, 47, 2008
|
|
6IO0
 
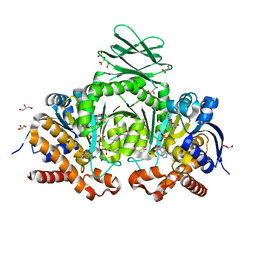 | | Human IDH1 R132C mutant complexed with compound A. | | Descriptor: | (2E)-3-{3-[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazole-4-carbonyl]-1-methyl-1H-indol-7-yl}prop-2-enoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Suzuki, M, Baba, D, Hanzawa, H. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Potent Blood-Brain Barrier-Permeable Mutant IDH1 Inhibitor Suppresses the Growth of Glioblastoma with IDH1 Mutation in a Patient-Derived Orthotopic Xenograft Model.
Mol.Cancer Ther., 19, 2020
|
|
6JMF
 
 | |
11AS
 
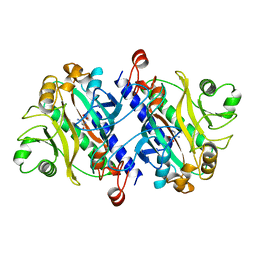 | |
12AS
 
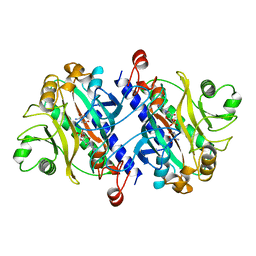 | | ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, ASPARAGINE SYNTHETASE | | Authors: | Nakatsu, T, Kato, H, Oda, J. | | Deposit date: | 1997-12-02 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
Nat.Struct.Biol., 5, 1998
|
|
2N42
 
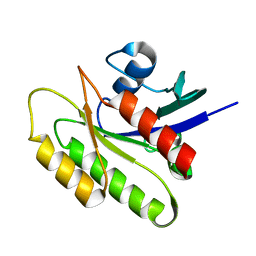 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N46
 
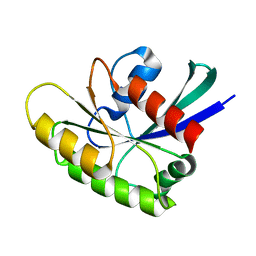 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
8K7P
 
 | | Staphylococcus aureus lipase -PSA complex | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Staphylococcus aureus lipase complex with unsaturated petroselinic acid.
Febs Open Bio, 14, 2024
|
|
8K7Q
 
 | | Staphylococcus aureus lipase S116A inactive mutant-PSA complex | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Staphylococcus aureus lipase complex with unsaturated petroselinic acid.
Febs Open Bio, 14, 2024
|
|
