3VNE
 
 | | Structure of the ebolavirus protein VP24 from Sudan | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
7WIJ
 
 | |
8H3D
 
 | | Structure of apo SARS-CoV-2 spike protein with one RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8H3E
 
 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8XAR
 
 | | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with SAM and Compound 54 | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-2-ethyl-5-pyridin-3-yl-pyrazolo[3,4-c]quinolin-4-one, CHLORIDE ION, ... | | Authors: | Zheng, J.Y, Zhang, G.P, Li, J.J, Tong, S.L. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with High Selectivity, Brain Penetration, and In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
7Y54
 
 | |
7Y6O
 
 | |
7Y4X
 
 | |
7Y5J
 
 | |
7Y5R
 
 | |
7Y73
 
 | |
7Y6E
 
 | |
7Y8H
 
 | |
7Y8S
 
 | |
7Y8I
 
 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y95
 
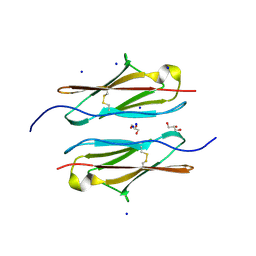 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | Descriptor: | Dscam, GLYCEROL, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y9A
 
 | | Crystal structure of sDscam Ig1-2 domains, isoform beta2v6 | | Descriptor: | Down Syndrome Cell Adhesion Molecules, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)][beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
8J40
 
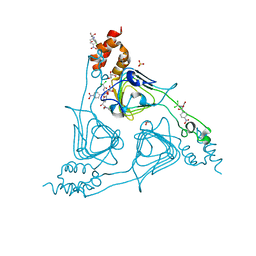 | | Crystal Structure of CATB8 in complex with chloramphenicol | | Descriptor: | CHLORAMPHENICOL, CatB8, GLYCEROL, ... | | Authors: | Liao, J, Kuang, L, Jiang, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Chloramphenicol Binding Sites of Acinetobacter baumannii Chloramphenicol Acetyltransferase CatB8.
Acs Infect Dis., 10, 2024
|
|
7CRN
 
 | |
8KHG
 
 | | Itaconyl-CoA hydratase PaIch | | Descriptor: | CITRIC ACID, Itaconyl-CoA hydratase | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8KHL
 
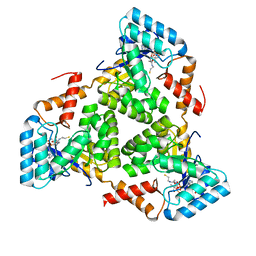 | | (S)-citramalyl-CoA lyase | | Descriptor: | COENZYME A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8K60
 
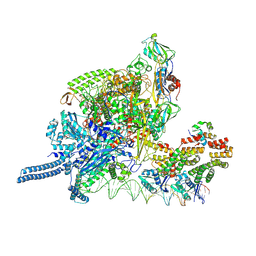 | |
7DXO
 
 | |
5Z62
 
 | | Structure of human cytochrome c oxidase | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, COPPER (II) ION, ... | | Authors: | Gu, J, Zong, S, Wu, M, Yang, M. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-27 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the intact 14-subunit human cytochrome c oxidase.
Cell Res., 28, 2018
|
|
6ADR
 
 | |
