4UQB
 
 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum at 1.68 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBOHYDRATE BINDING FAMILY 6, SULFATE ION | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-06-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4UCI
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | ADENOSINE, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
4UCV
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UHQ
 
 | | Crystal structure of the pyocin AP41 DNase | | Descriptor: | CITRIC ACID, LARGE COMPONENT OF PYOCIN AP41, NICKEL (II) ION | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
4UJC
 
 | | mammalian 80S HCV-IRES initiation complex with eIF5B POST-like state | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Initiation Factor 5B on Hcv-Ires RNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UOB
 
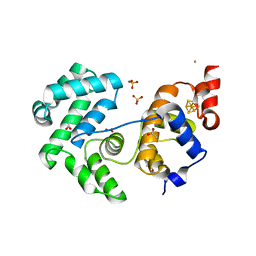 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | Descriptor: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
7F4X
 
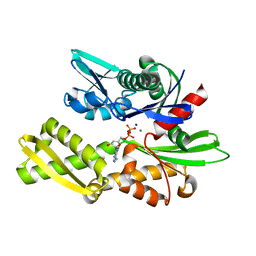 | | Joint neutron and X-ray crystal structure of the nucleotide-binding domain of Hsp72 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7FGX
 
 | | Toxoplasma gondii dihydrofolate reductase thymidylate synthase (TgDHFR-TS) complexed with P39, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6-hexyl-5-phenyl-pyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Yoomuang, A, Taweechai, S, Saeyang, T, Yuvaniyama, J, Tarnchompoo, B, Yuthavong, Y, Kamchonwongpaisan, S. | | Deposit date: | 2021-07-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into Effective Inhibitors' Binding to Toxoplasma gondii Dihydrofolate Reductase Thymidylate Synthase.
Acs Chem.Biol., 17, 2022
|
|
7FGY
 
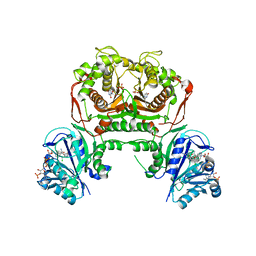 | | Toxoplasma gondii dihydrofolate reductase thymidylate synthase (TgDHFR-TS) complexed with P40, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(3-chlorophenyl)-6-[2-(3-phenoxypropoxy)ethyl]pyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Yoomuang, A, Taweechai, S, Saeyang, T, Yuvaniyama, J, Tarnchompoo, B, Yuthavong, Y, Kamchonwongpaisan, S. | | Deposit date: | 2021-07-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Insight into Effective Inhibitors' Binding to Toxoplasma gondii Dihydrofolate Reductase Thymidylate Synthase.
Acs Chem.Biol., 17, 2022
|
|
4UN5
 
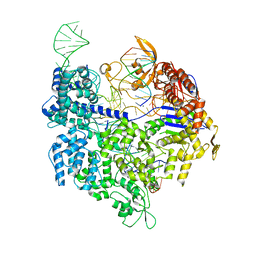 | | Crystal structure of Cas9 bound to PAM-containing DNA target containing mismatches at positions 1-3 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN4
 
 | | Crystal structure of Cas9 bound to PAM-containing DNA target with mismatches at positions 1-2 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
7FGW
 
 | | Toxoplasma gondii dihydrofolate reductase thymidylate synthase (TgDHFR-TS) complexed with pyrimethamine, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, BENZAMIDINE, ... | | Authors: | Vanichtanankul, J, Yoomuang, A, Taweechai, S, Saeyang, T, Yuvaniyama, Y, Tarnchompoo, B, Yuthavong, Y, Kamchonwongpaisan, S. | | Deposit date: | 2021-07-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insight into Effective Inhibitors' Binding to Toxoplasma gondii Dihydrofolate Reductase Thymidylate Synthase.
Acs Chem.Biol., 17, 2022
|
|
7F7A
 
 | | Crystal structure of Non-specific class-C acid phosphatase from Sphingobium sp. RSMS bound to Adenine at pH 9 | | Descriptor: | ADENINE, Acid phosphatase, MAGNESIUM ION | | Authors: | Gaur, N.K, Kumar, A, Sunder, S, Mukhopadhyaya, R, Makde, R.D. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS
To Be Published
|
|
7F7D
 
 | | Crystal structure of Non-specific class-C acid phosphatase from Sphingobium sp. RSMS bound to Adenosine at pH 5.5 | | Descriptor: | ADENOSINE, Acid phosphatase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gaur, N.K, Kumar, A, Sunder, S, Mukhopadhyaya, R, Makde, R.D. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS
To Be Published
|
|
4UQC
 
 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum at 1.30 A resolution | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-06-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7F80
 
 | |
7F9U
 
 | |
4UJ8
 
 | | Structure of surface layer protein SbsC, domains 6-7 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Surface Layer Protein Sbsc
To be Published
|
|
7F9R
 
 | | Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in Complex with inhibitor L95, L-Proline and Febrifugine | | Descriptor: | 1,2-ETHANEDIOL, 3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, CHLORIDE ION, ... | | Authors: | Manickam, Y, Malhotra, N, Sharma, A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | TgPRS with double inhibitors
To Be Published
|
|
7F7B
 
 | | Crystal structure of Non-specific class-C acid phosphatase from Sphingobium sp. RSMS bound to BIS-TRIS at pH 5.5 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acid phosphatase, MAGNESIUM ION, ... | | Authors: | Gaur, N.K, Kumar, A, Sunder, S, Mukhopadhyaya, R, Makde, R.D. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS
To Be Published
|
|
7F9V
 
 | |
7F9T
 
 | |
4UCE
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M72 | | Descriptor: | 1-[(4-fluorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UG1
 
 | | GpsB N-terminal domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, IMIDAZOLE, NICKEL (II) ION | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Muller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
4UCC
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M76 | | Descriptor: | 1-[(2,4-dichlorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
