4F2N
 
 | |
7K0F
 
 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | Descriptor: | 3C-like proteinase, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ZAN
 
 | |
7JPM
 
 | |
8QXL
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State II - Hemi-relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXJ
 
 | | Cryo-EM structure of tetrameric human SAMHD1 with dApNHpp | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXM
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State III - Relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXN
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State IV - Depleted relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXK
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State I - Tense | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8QXO
 
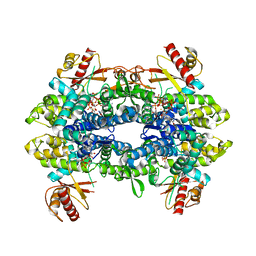 | | Cryo-EM structure of tetrameric human SAMHD1 State V - Depleted relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
6CQA
 
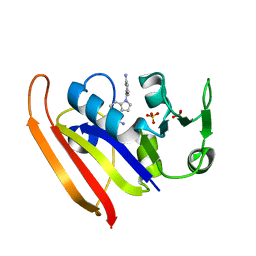 | | E. coli DHFR complex with inhibitor AMPQD | | Descriptor: | 7-[(3-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cao, H, Rodrigues, J, Benach, J, Wasserman, S, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
5OAC
 
 | |
7EQE
 
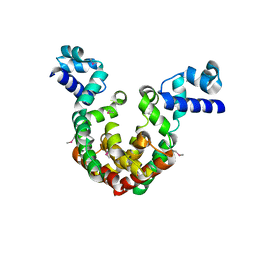 | | Crystal Structure of a transcription factor | | Descriptor: | TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2022-10-19 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
7EQF
 
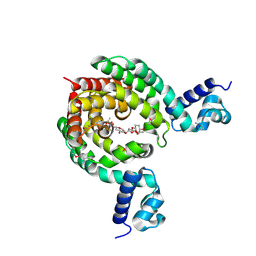 | | Crystal Structure of a Transcription Factor in complex with Ligand | | Descriptor: | (6~{R})-3-methyl-8-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{R},5~{R},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{S},5~{S},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-oxidanyl-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-1,6,11-tris(oxidanyl)-5,6-dihydrobenzo[a]anthracene-7,12-dione, TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
5VAA
 
 | | Crystal structure of mouse IgG2a Fc T370K mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Ig gamma-2A chain C region, ... | | Authors: | Armstrong, A.A, Gilliland, G.L. | | Deposit date: | 2017-03-24 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Efficient Generation of Bispecific Murine Antibodies for Pre-Clinical Investigations in Syngeneic Rodent Models.
Sci Rep, 7, 2017
|
|
8T36
 
 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
8T4T
 
 | |
8T31
 
 | |
8T35
 
 | | Crystal structure of K51 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
8T33
 
 | | Crystal structure of K46 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | ACETATE ION, Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2, ... | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
8T32
 
 | | Crystal structure of K48 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, LIR of DOR, TRIETHYLENE GLYCOL | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
3ZG1
 
 | | NI-BOUND FORM OF M123A MUTANT OF CUPRIAVIDUS METALLIDURANS CH34 CNRXS | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Girard, E, Coves, J. | | Deposit date: | 2012-12-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal Sensing and Signal Transduction by Cnrx from Cupriavidus Metallidurans Ch34: Role of the Only Methionine Assessed by a Functional, Spectroscopic, and Theoretical Study
Metallomics, 6, 2014
|
|
6D7J
 
 | |
