6VFC
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min)
To be published
|
|
1PM9
 
 | | CRYSTAL STRUCTURE OF HUMAN MNSOD H30N, Y166F MUTANT | | Descriptor: | MANGANESE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
6BZP
 
 | | STGGYG from low-complexity domain of FUS, residues 77-82 | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, RNA-binding protein FUS | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6VS9
 
 | |
1PKJ
 
 | | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional deaminase/diphosphatase, DEOXYURIDINE-5'-TRIPHOSPHATE | | Authors: | Huffman, J.L, Li, H, White, R.H, Tainer, J.A. | | Deposit date: | 2003-06-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii
J.Mol.Biol., 331, 2003
|
|
1PL4
 
 | | Crystal Structure of human MnSOD Y166F mutant | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
3F5A
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 3'SiaLacNAc1-3 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
1PW6
 
 | | Low Micromolar Small Molecule Inhibitor of IL-2 | | Descriptor: | 2-CYCLOHEXYL-N-(2-{4-[5-(2,3-DICHLORO-PHENYL)-2H-PYRAZOL-3-YL]-PIPERIDIN-1-YL}-2-OXO-ETHYL)-2-GUANIDINO-ACETAMIDE, Interleukin-2, SULFATE ION | | Authors: | Thanos, C.D, Randal, M, Wells, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-01-13 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent small-molecule binding to a dynamic hot spot on IL-2.
J.Am.Chem.Soc., 125, 2003
|
|
1Q2Z
 
 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
3FK4
 
 | | Crystal structure of RuBisCO-like protein from Bacillus Cereus ATCC 14579 | | Descriptor: | RuBisCO-like protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-15 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of RuBisCO-like protein from Bacillus Cereus ATCC 14579
To be Published
|
|
6VV7
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB136 | | Descriptor: | 5-[3-(5-methyl-1H-indol-3-yl)propoxy]-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B, Chavez-Pacheco, S.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
1PKK
 
 | | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Bifunctional deaminase/diphosphatase | | Authors: | Huffman, J.L, Li, H, White, R.H, Tainer, J.A. | | Deposit date: | 2003-06-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii
J.Mol.Biol., 331, 2003
|
|
3F53
 
 | |
3G0D
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 2 | | Descriptor: | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
3G1X
 
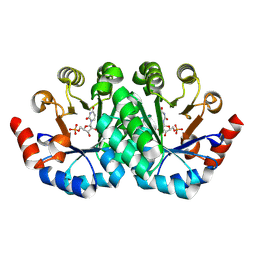 | | Crystal structure of the mutant D70G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
6BYD
 
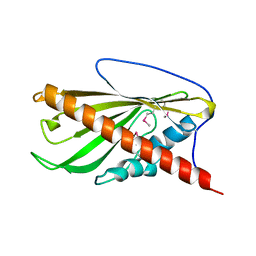 | | Crystal structure of the second StART domain of yeast Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
1NIW
 
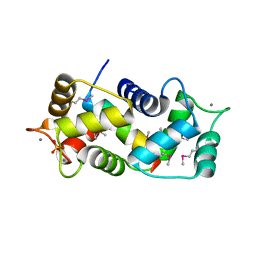 | | Crystal structure of endothelial nitric oxide synthase peptide bound to calmodulin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nitric-oxide synthase, ... | | Authors: | Aoyagi, M, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-12-26 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for endothelial nitric oxide synthase binding to calmodulin
Embo J., 22, 2003
|
|
6VSD
 
 | |
6VSG
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(trifluoromethyl)benzene-1,2-diamine(fragment 17) | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
3G1D
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
1NBQ
 
 | | Crystal Structure of Human Junctional Adhesion Molecule Type 1 | | Descriptor: | Junctional adhesion molecule 1 | | Authors: | Prota, A.E, Campbell, J.A, Schelling, P, Forrest, J.C, Watson, M.J, Peters, T.R, Aurrand-Lions, M, Imhof, B.A, Dermody, T.S, Stehle, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6CLC
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.27 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLH
 
 | | 1.37 A MicroED structure of GSNQNNF at 2.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.37 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLP
 
 | | 1.16 A MicroED structure of GSNQNNF at 2.5 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.16 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
