8K3L
 
 | | SOD1 and Nanobody3 complex | | Descriptor: | COPPER (II) ION, NB3, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cheng, S, Liu, R, Ding, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SOD1 and Nanobody1 complex
To Be Published
|
|
2I7O
 
 | | Structure of Re(4,7-dimethyl-phen)(Thr124His)(Lys122Trp)(His83Gln)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Sudhamsu, J, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tryptophan-accelerated electron flow through proteins.
Science, 320, 2008
|
|
3WQ9
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 2-(4-Hydroxy-cyclohexylamino)-4-[5-(4-phenyl-imidazol-1-yl)-isoquinolin-1-yl]-benzamide | | Descriptor: | 2-[(trans-4-hydroxycyclohexyl)amino]-4-[5-(4-phenyl-1H-imidazol-1-yl)isoquinolin-1-yl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Chong, K.T, Yamashita, S, Oshiumi, H, Uno, T, Kitade, M. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of highly selective Hsp90 / inhibitors by structure and thermodynamics guided design
To be Published
|
|
5JA1
 
 | | EntF, a Terminal Nonribosomal Peptide Synthetase Module Bound to the MbtH-Like Protein YbdZ | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, CHLORIDE ION, Enterobactin biosynthesis protein YbdZ, ... | | Authors: | Miller, B.R, Gulick, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
J.Biol.Chem., 291, 2016
|
|
6PAK
 
 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Subtilisin E | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-06-11 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Enhancing subtilisin thermostability through a modified normalized B-factor analysis and loop-grafting strategy.
J.Biol.Chem., 294, 2019
|
|
5X57
 
 | | Structure of GAR domain of ACF7 | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, NICKEL (II) ION | | Authors: | Yang, F, Wang, T, Zhang, Y, Wu, X.Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics.
Nat Commun, 8, 2017
|
|
5JA2
 
 | | EntF, a Terminal Nonribosomal Peptide Synthetase Module Bound to the non-Native MbtH-Like Protein PA2412 | | Descriptor: | 5'-({[(2R,3S)-3-amino-4-hydroxy-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}butyl]sulfonyl}amino)-5'-deoxyadenosine, Enterobactin synthase component F, MbtH-Like Protein PA2412, ... | | Authors: | Miller, B.R, Gulick, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a Nonribosomal Peptide Synthetase Module Bound to MbtH-like Proteins Support a Highly Dynamic Domain Architecture.
J.Biol.Chem., 291, 2016
|
|
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6T9O
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6UZ3
 
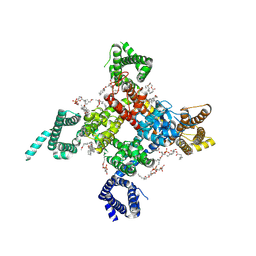 | | Cardiac sodium channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
6UZ0
 
 | | Cardiac sodium channel with flecainide | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
3DJ3
 
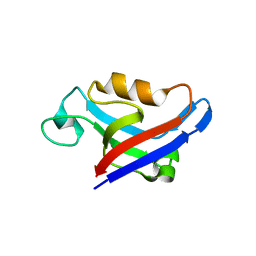 | |
3DIW
 
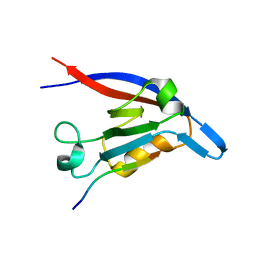 | | c-terminal beta-catenin bound TIP-1 structure | | Descriptor: | Tax1-binding protein 3, decameric peptide form Catenin beta-1 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
3DJ1
 
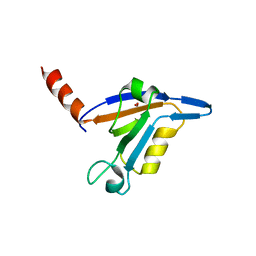 | | crystal structure of TIP-1 wild type | | Descriptor: | SULFATE ION, Tax1-binding protein 3 | | Authors: | Shen, Y. | | Deposit date: | 2008-06-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of beta-Catenin Recognition by Tax-interacting Protein-1
J.Mol.Biol., 384, 2008
|
|
8WPU
 
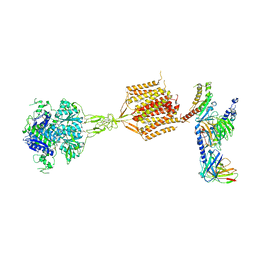 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPG
 
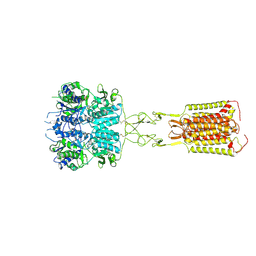 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
7CRF
 
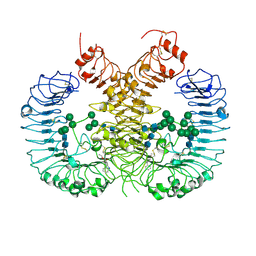 | | Crystal structure of human TLR8 in complex with CU-CPD107 | | Descriptor: | 1-[2-(ethoxymethyl)-4-iodanyl-5-phenyl-imidazol-1-yl]-2-methyl-propan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Sakaniwa, K, Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2020-08-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Tetrasubstituted imidazoles as incognito Toll-like receptor 8 a(nta)gonists.
Nat Commun, 12, 2021
|
|
8GUN
 
 | |
8GUO
 
 | |
8GUP
 
 | | Crystal structure of EsaG from Staphylococcus aureus | | Descriptor: | CITRIC ACID, Type VII secretion system protein EsaG | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.298725 Å) | | Cite: | A toxin-deformation dependent inhibition mechanism in the T7SS toxin-antitoxin system of Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
4UMX
 
 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | 2,6-bis(1H-imidazol-1-ylmethyl)-4-(2,4,4-trimethylpentan-2-yl)phenol, GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, ... | | Authors: | Mathieu, M, Marquette, J.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
4UMY
 
 | | IDH1 R132H in complex with cpd 1 | | Descriptor: | GLYCEROL, ISOCITRATE DEHYDROGENASE [NADP] CYTOPLASMIC, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McLean, L, Zhang, Y, Mathieu, M. | | Deposit date: | 2014-05-22 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Selective Inhibition of Mutant Isocitrate Dehydrogenase 1 (Idh1) Via Disruption of a Metal Binding Network by an Allosteric Small Molecule.
J.Biol.Chem., 290, 2015
|
|
7DTW
 
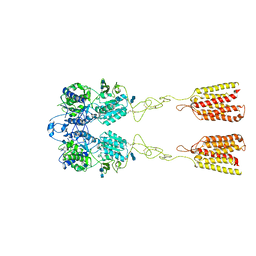 | | Human Calcium-Sensing Receptor in the inactive close-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTU
 
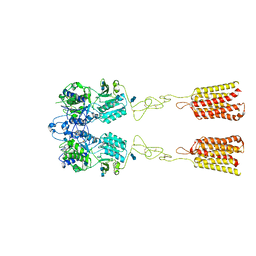 | | Human Calcium-Sensing Receptor bound with L-Trp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Extracellular calcium-sensing receptor, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Liu, L, Sun, D.M, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
7DTT
 
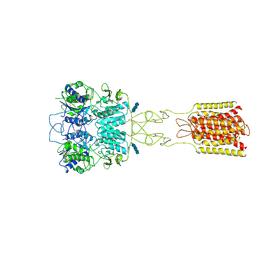 | | Human Calcium-Sensing Receptor bound with calcium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ling, S.L, Tian, C.L, Shi, P, Liu, S.L, Meng, X.Y, Sun, D.M, Liu, L, Shi, C.W. | | Deposit date: | 2021-01-06 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of cooperative activation of the human calcium-sensing receptor by Ca 2+ ions and L-tryptophan.
Cell Res., 31, 2021
|
|
