9EX1
 
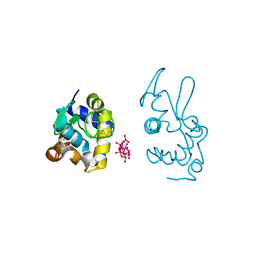 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure B) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4PPO
 
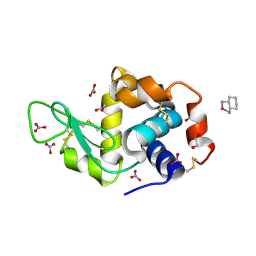 | | First Crystal Structure for an Oxaliplatin-Protein Complex | | Descriptor: | 1,2-ETHANEDIOL, CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), Lysozyme C, ... | | Authors: | Messori, L, Merlino, A. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The X-ray structure of the complex formed in the reaction between oxaliplatin and lysozyme.
Chem.Commun.(Camb.), 50, 2014
|
|
9EX2
 
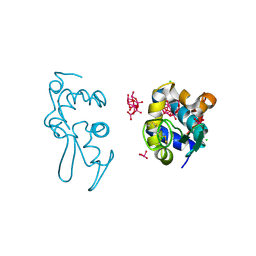 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure C) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4QY9
 
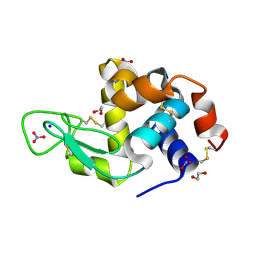 | | X-ray structure of the adduct between hen egg white lysozyme and Auoxo3, a cytotoxic gold(III) compound | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Lysozyme C, ... | | Authors: | Russo Krauss, I, Merlino, A. | | Deposit date: | 2014-07-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Interactions of gold-based drugs with proteins: the structure and stability of the adduct formed in the reaction between lysozyme and the cytotoxic gold(iii) compound Auoxo3.
Dalton Trans, 43, 2014
|
|
8BRC
 
 | | Crystal structure of the adduct between human serum transferrin and cisplatin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIA, FE (III) ION, ... | | Authors: | Troisi, R, Galardo, F, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Cisplatin Binding to Human Serum Transferrin: A Crystallographic Study.
Inorg.Chem., 62, 2023
|
|
9H49
 
 | | Crystal structure of the adduct between human serum transferrin (apo-form) and cisplatin | | Descriptor: | AMMONIA, CITRIC ACID, PLATINUM (II) ION, ... | | Authors: | Troisi, R, Galardo, F, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2024-10-17 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Cisplatin/Apo-Transferrin Adduct: X-ray Structure and Binding to the Transferrin Receptor 1.
Inorg.Chem., 64, 2025
|
|
5O7Q
 
 | | Crystal structure of a single chain monellin mutant (Y65R) pH 5.5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Pica, A, Merlino, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | pH driven fibrillar aggregation of the super-sweet protein Y65R-MNEI: A step-by-step structural analysis.
Biochim. Biophys. Acta, 1862, 2017
|
|
8PTE
 
 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to octadecavanadate ion (structure B) | | Descriptor: | Lysozyme C, NITRATE ION, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Ferraro, G, Merlino, A. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4NY5
 
 | |
5O7L
 
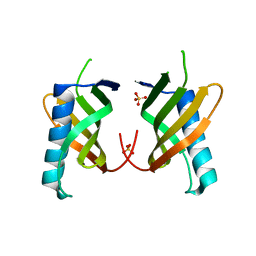 | |
5O7S
 
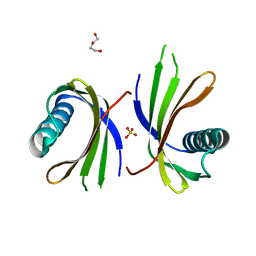 | | Crystal structure of a single chain monellin mutant (Y65R) pH 8.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Pica, A, Merlino, A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | pH driven fibrillar aggregation of the super-sweet protein Y65R-MNEI: A step-by-step structural analysis.
Biochim. Biophys. Acta, 1862, 2017
|
|
5O7R
 
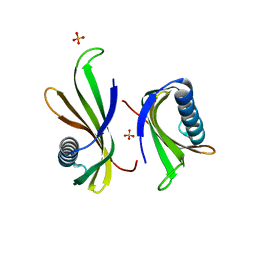 | |
5O7K
 
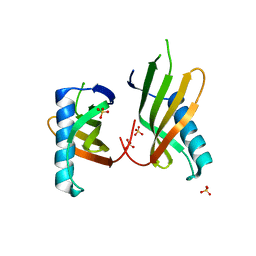 | |
9GOG
 
 | | X-ray structure of lysozyme obtained upon reaction with the dioxidovanadium(V) complex with furan-2-carboxylic acid (3-ethoxy-2-hydroxybenzylidene)hydrazide | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Paolillo, M, Ferraro, G, Merlino, A. | | Deposit date: | 2024-09-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Interaction of V V O 2 -hydrazonates with lysozyme.
J.Inorg.Biochem., 264, 2024
|
|
9GOK
 
 | | X-ray structure of lysozyme obtained upon reaction with the dioxidovanadium(V) complex with (E)-N'-(1-(2-hydroxy-5-methoxyphenyl)ethylidene)furan-2-carbohydrazide) | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Paolillo, M, Ferraro, G, Merlino, A. | | Deposit date: | 2024-09-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Interaction of V V O 2 -hydrazonates with lysozyme.
J.Inorg.Biochem., 264, 2024
|
|
5NA9
 
 | |
9HMQ
 
 | | X-structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with lysozyme (structure C) | | Descriptor: | AMMONIA, IODIDE ION, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
9I8L
 
 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] at 310 K | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Ferraro, G, Merlino, A. | | Deposit date: | 2025-02-05 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Formation of Mixed-Valence Cage-Like Polyoxidovanadates at 37°C Upon Reaction of V IV O(acetylacetonato) 2 With Lysozyme.
Chemistry, 31, 2025
|
|
9HMK
 
 | | X-ray structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with lysozyme (structure B) | | Descriptor: | IODIDE ION, Lysozyme C, PLATINUM (II) ION | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-09 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
9HN6
 
 | | X-ray structure of the adduct formed upon reaction of the diiodido analogue of picoplatin with ribonuclease A | | Descriptor: | AMMONIA, CHLORIDE ION, IODIDE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cytotoxicity and Binding to DNA, Lysozyme, Ribonuclease A, and Human Serum Albumin of the Diiodido Analog of Picoplatin.
Inorg.Chem., 64, 2025
|
|
9HNB
 
 | |
9H4V
 
 | | Crystal structure of the adduct formed upon reaction of aurothiomalate with human serum transferrin (apo-form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, GOLD ION, ... | | Authors: | Troisi, R, Galardo, F, Messori, L, Sica, F, Merlino, A. | | Deposit date: | 2024-10-21 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The X-ray structure of the adduct formed upon reaction of aurothiomalate with apo-transferrin: gold binding sites and a unique transferrin structure along the apo/holo transition pathway
Inorg Chem Front, 12, 2025
|
|
5MIJ
 
 | | X-ray structure of carboplatin-encapsulated horse spleen apoferritin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Pontillo, N, Ferraro, G, Helliwell, J.R, Merlino, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure of the Carboplatin-Loaded Apo-Ferritin Nanocage.
ACS Med Chem Lett, 8, 2017
|
|
9FMY
 
 | |
5NJ1
 
 | |
