6LD2
 
 | | Zika NS5 polymerase domain | | Descriptor: | (1S,2S,4S,5R)-2,4-dimethoxy-5-thiophen-2-yl-cyclohexane-1-carboxylic acid, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
8T9D
 
 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
6LD1
 
 | | Zika NS5 polymerase domain | | Descriptor: | 1,2-ETHANEDIOL, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
8FGX
 
 | | Cryo-EM structure of the STAR-0215 Fab in complex with active human plasma kallikrein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma kallikrein light chain, STAR-0215 Heavy chain, ... | | Authors: | Fuller, J.R, Biris, N, Bista, P. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | STAR-0215 is a Novel, Long-Acting Monoclonal Antibody Inhibitor of Plasma Kallikrein for the Potential Treatment of Hereditary Angioedema.
J.Pharmacol.Exp.Ther., 387, 2023
|
|
7FJP
 
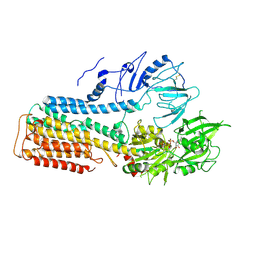 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7FJQ
 
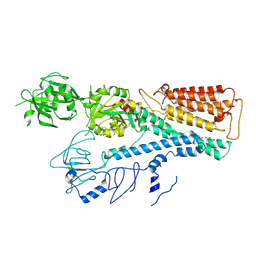 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | Polyamine-transporting ATPase 13A2, SPERMINE | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
6LD5
 
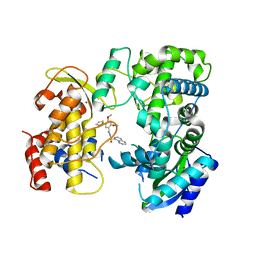 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, RNA-directed RNA polymerase NS5, ZINC ION, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
6LD4
 
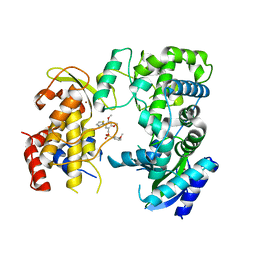 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, 3-methoxybenzenesulfonamide, RNA-directed RNA polymerase NS5, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
8TYP
 
 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
6LD3
 
 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, RNA-directed RNA polymerase NS5, ZINC ION, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
7MKM
 
 | |
6L4Q
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-B | | Descriptor: | (3R)-3-[[(3R)-3-methylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y, Mishra, S, Harlos, K. | | Deposit date: | 2019-10-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
6L3Y
 
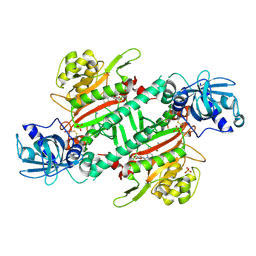 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Clado-C | | Descriptor: | (3R)-3-[[(3S)-3-ethylpiperidin-1-yl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Babbar, P, Sharma, A, Mishra, S, Manickam, Y, Harlos, K. | | Deposit date: | 2019-10-15 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin inhibitor, Cla-B
Chembiochem, 2021
|
|
7MKL
 
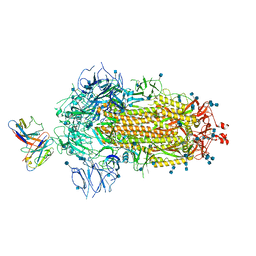 | |
5K5E
 
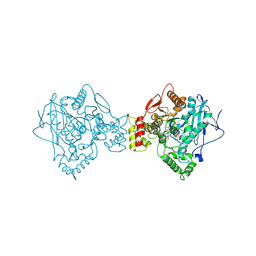 | | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | De la Mora, E, Dighe, S.N, Deora, G.S, Ross, B.P, Nachon, F, Brazzolotto, X. | | Deposit date: | 2016-05-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening.
J.Med.Chem., 59, 2016
|
|
6EZ2
 
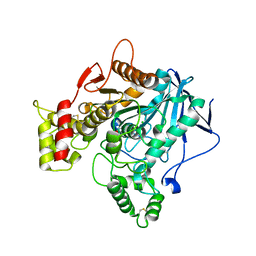 | | Human butyrylcholinesterase carbamylated. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, de la Mora, E, Dighe, S.N, Ross, B.P. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rivastigmine and metabolite analogues with putative Alzheimer's disease-modifying properties in a Caenorhabditis elegans model.
Commun Chem, 2019
|
|
7DMX
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | Descriptor: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7WMH
 
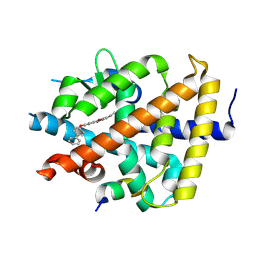 | | A novel chemical derivative(56) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenylphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMG
 
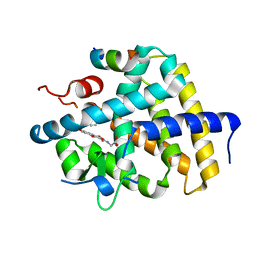 | | A novel chemical derivative(52) of THRB agonist | | Descriptor: | 2-[[1-methoxy-4-oxidanyl-7-(4-phenoxyphenoxy)isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7WMJ
 
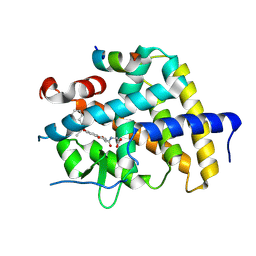 | | A novel chemical derivative(71) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(phenylcarbonyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
6ZZB
 
 | |
7DNA
 
 | |
