7XHN
 
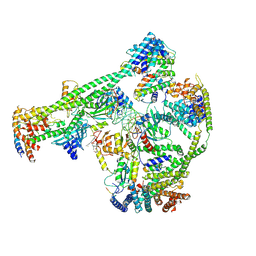 | | Structure of human inner kinetochore CCAN-DNA complex | | Descriptor: | CENP-W, Centromere protein C, Centromere protein H, ... | | Authors: | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | Deposit date: | 2022-04-09 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
7XDQ
 
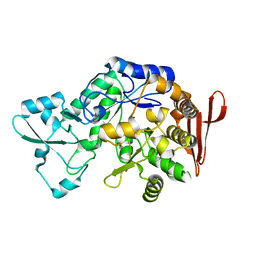 | | Crystal structure of a glucosylglycerol phosphorylase mutant from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase, LITHIUM ION, beta-D-glucopyranose | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7XDR
 
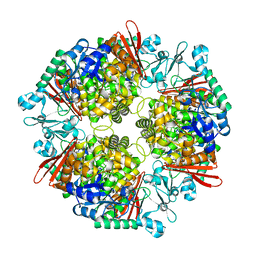 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
4P3K
 
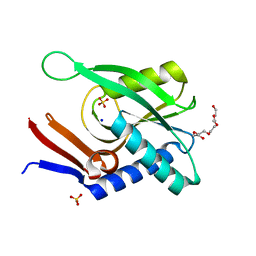 | | Structure of ancestral PyrR protein (PLUMPyrR) | | Descriptor: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P47
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM OCHROBACTRUM ANTHROPI (Oant_4429), TARGET EFI-510151, C-TERMIUS BOUND IN LIGAND BINDING POCKET | | Descriptor: | TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-12 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PH2
 
 | | mature N-terminal domain of capsid protein from bovine leukemia virus | | Descriptor: | BLV capsid - N-terminal domain, GLYCEROL, SULFATE ION | | Authors: | Trajtenberg, F, Obal, G, Pritsch, O, Buschiazzo, A. | | Deposit date: | 2014-05-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | STRUCTURAL VIROLOGY. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography.
Science, 349, 2015
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
4PFI
 
 | | Crystal structure of a tRAP periplasmic solute binding protein from marinobacter aquaeolei VT8 (Maqu_2829, TARGET EFI-510133), apo open structure | | Descriptor: | SODIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3024 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PCD
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS OCh 114 (RD1_1052, TARGET EFI-510238) WITH BOUND L-GALACTONATE | | Descriptor: | C4-dicarboxylate transport system, substrate-binding protein, putative, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PAK
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM VERMINEPHROBACTER EISENIAE EF01-2 (Veis_3954, TARGET EFI-510324) A NEPHRIDIAL SYMBIONT OF THE EARTHWORM EISENIA FOETIDA, BOUND TO (R)-PANTOIC ACID | | Descriptor: | PANTOATE, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
7Y9J
 
 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium in complex with 5-nitro-1,2-benzisoxazole | | Descriptor: | 5-nitro-1,2-benzoxazole, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
7Y9K
 
 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
4RF5
 
 | | Crystal structure of ketoreductase from Lactobacillus kefir, E145S mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | Authors: | Tang, Y, Tibrewal, N, Cascio, D. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7X8X
 
 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
4RF3
 
 | | Crystal Structure of ketoreductase from Lactobacillus kefir, mutant A94F | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | Authors: | Tang, Y, Tibrewal, N, Cascio, D. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4P3L
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CHROMOHALOBACTER SALEXIGENS DSM 3043 (Csal_2479), TARGET EFI-510085, WITH BOUND GLUCURONATE, SPG P6122 | | Descriptor: | CHLORIDE ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-09 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P1L
 
 | | Crystal structure of a trap periplasmic solute binding protein from chromohalobacter salexigens dsm 3043 (csal_2479), target EFI-510085, with bound d-glucuronate, spg i213 | | Descriptor: | GLYCEROL, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P1E
 
 | | Crystal structure of a trap periplasmic solute binding protein from escherichia fergusonii (efer_1530), target EFI-510119, apo open structure, phased with iodide | | Descriptor: | IODIDE ION, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
7VOA
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
4PDD
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM POLAROMONAS SP JS666 (Bpro_0088, TARGET EFI-510167) BOUND TO D-ERYTHRONATE | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, FORMIC ACID, MALONIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PF8
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1 (TARGET EFI-510299) WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP-T family transporter, DctP (Periplasmic binding) subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PH0
 
 | |
4RPA
 
 | |
4P80
 
 | | Structure of ancestral PyrR protein (AncGREENPyrR) | | Descriptor: | Ancestral PyrR protein (Green), SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
