4J9I
 
 | |
4J9F
 
 | |
4J9E
 
 | |
4J9B
 
 | |
4J9D
 
 | |
4JJB
 
 | |
4J9H
 
 | |
4J9C
 
 | |
4J9G
 
 | |
4JJD
 
 | |
7KLM
 
 | | Human Arginase1 Complexed with Inhibitor Compound 24a | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLL
 
 | | Human Arginase1 Complexed with Inhibitor Compound 18 | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-azanyl-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLK
 
 | | Human Arginase1 Complexed with Inhibitor Compound 3a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-carboxypiperidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
3OB6
 
 | | Structure of AdiC(N101A) in the open-to-out Arg+ bound conformation | | Descriptor: | ARGININE, AdiC arginine:agmatine antiporter | | Authors: | Carpena, X, Kowalczyk, L, Ratera, M, Valencia, E, Vazquez-lbar, J.L, Fita, I, Palacin, M. | | Deposit date: | 2010-08-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate-induced permeation by an amino acid antiporter.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5HLP
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
5HLN
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH CHIR99021 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHIR99021, Glycogen synthase kinase-3 beta, ... | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
6YQ4
 
 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
6C6I
 
 | | Crystal structure of a chimeric NDM-1 metallo-beta-lactamase harboring the IMP-1 L3 loop | | Descriptor: | Metallo-beta-lactamase type 2 chimera, ZINC ION | | Authors: | Otero, L, Giannini, E, Klinke, S, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
7TV9
 
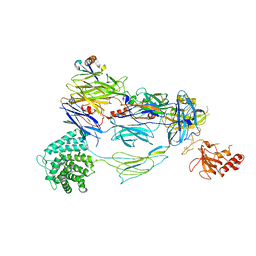 | | HUMAN COMPLEMENT COMPONENT C3B IN COMPLEX WITH APL-1030 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APL-1030 Nanofitin, Complement C3 beta chain, ... | | Authors: | Fontano, E, Nadupalli, A, Lakshminarasimhan, D, White, A, Garlish, J, Cinier, M, Chevrel, A, Perrocheau, A, Eyerman, D, Orme, M, Kitten, O, Scheibler, L. | | Deposit date: | 2022-02-04 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of APL-1030, a Novel, High-Affinity Nanofitin Inhibitor of C3-Mediated Complement Activation.
Biomolecules, 12, 2022
|
|
6CAC
 
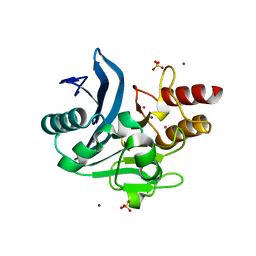 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
8BT9
 
 | |
6VNS
 
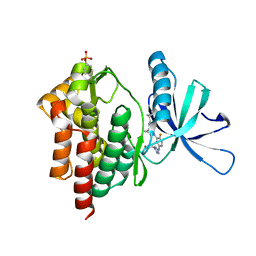 | | Crystal structure of TYK2 kinase with compound 13 | | Descriptor: | (1R,2R)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNY
 
 | | Crystal structure of TYK2 kinase with compound 10 | | Descriptor: | N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNX
 
 | | Crystal structure of TYK2 kinase with compound 19 | | Descriptor: | (1S)-2,2-difluoro-N-[(1S,5R,6R)-3-{5-fluoro-2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-6-methyl-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
6VNV
 
 | | Crystal structure of TYK2 kinase with compound 14 | | Descriptor: | (1S,2S)-2-cyano-N-[(1S,5R)-3-(5-fluoro-2-{[1-(2-hydroxyethyl)-1H-pyrazol-4-yl]amino}pyrimidin-4-yl)-3-azabicyclo[3.1.0]hexan-1-yl]cyclopropane-1-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and optimization of a series of 4-(3-azabicyclo[3.1.0]hexan-3-yl)pyrimidin-2-amines: Dual inhibitors of TYK2 and JAK1.
Bioorg.Med.Chem., 28, 2020
|
|
