4BIC
 
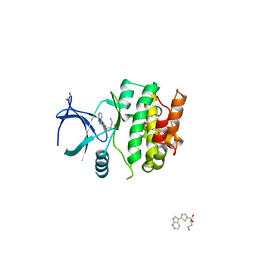 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4O22
 
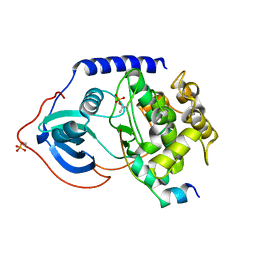 | | Binary complex of metal-free PKAc with SP20. | | Descriptor: | Phosphorylated peptide pSP20., cAMP-dependent protein kinase catalytic subunit alpha. | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S.S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
4BF2
 
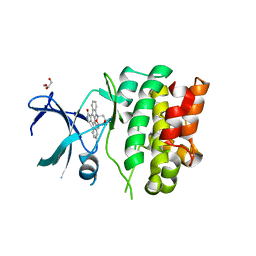 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | ACETATE ION, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BID
 
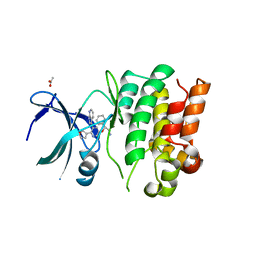 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 5-[(1R)-2-amino-1-phenylethoxy]-2-(furan-3-yl)-3-{1H-pyrrolo[2,3-b]pyridin-3-yl}pyridine, ACETATE ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
5NGJ
 
 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
4O21
 
 | | Product complex of metal-free PKAc, ATP-gamma-S and SP20. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thio-phosphorylated peptide pSP20, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
4BIE
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{2-methyl-1H-pyrrolo[2,3-b]pyridin-4-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIB
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 3-cyano-4-(piperidin-4-yloxy)-1H-indole-7-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4PDJ
 
 | | Neutron crystal Structure of E.coli Dihydrofolate Reductase complexed with folate and NADP+ | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Wan, Q, Kovalevsky, A.Y, Wilson, M, Langan, P, Dealwis, C, Bennett, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.599 Å), X-RAY DIFFRACTION | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1H70
 
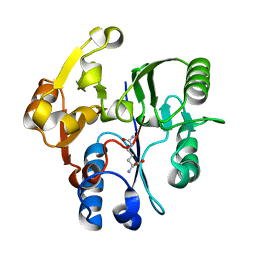 | | DDAH FROM PSEUDOMONAS AERUGINOSA. C249S MUTANT COMPLEXED WITH CITRULLINE | | Descriptor: | CITRULLINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE | | Authors: | Murray-Rust, J, Leiper, J, McAlister, M, Phelan, J, Tilley, S, Santamaria, J, Vallance, P, McDonald, N. | | Deposit date: | 2001-06-30 | | Release date: | 2001-08-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase.
Nat. Struct. Biol., 8, 2001
|
|
3QZA
 
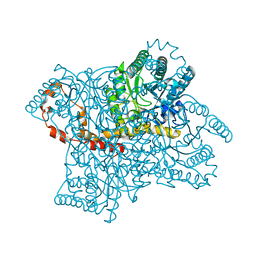 | |
3QBA
 
 | | Reintroducing Electrostatics into Macromolecular Crystallographic Refinement: Z-DNA (X-ray) | | Descriptor: | Z-DNA | | Authors: | Fenn, T.D, Schnieders, M.J, Mustyakimov, M, Wu, C, Langan, P, Pande, V.S, Brunger, A.T. | | Deposit date: | 2011-01-12 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.53 Å), X-RAY DIFFRACTION | | Cite: | Reintroducing electrostatics into macromolecular crystallographic refinement: application to neutron crystallography and DNA hydration.
Structure, 19, 2011
|
|
3QYS
 
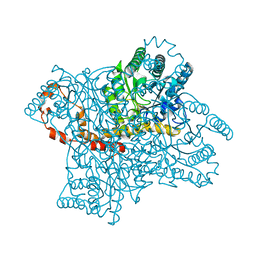 | |
4A56
 
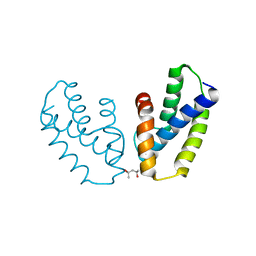 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | Authors: | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
4DH5
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ADP, Phosphate, and IP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH3
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DUO
 
 | | Room-temperature X-ray structure of D-Xylose Isomerase in complex with 2Mg2+ ions and xylitol at pH 7.7 | | Descriptor: | MAGNESIUM ION, Xylitol, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH7
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4QYT
 
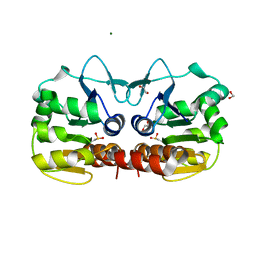 | | Schizosaccharomyces pombe DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein C22E12.03c | | Authors: | Wilson, M.A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Influence of peptide dipoles and hydrogen bonds on reactive cysteine pK(a) values in fission yeast DJ-1.
Febs J., 279, 2012
|
|
9FAC
 
 | | Additional cryo-EM structure of cardiac amyloid AL59 - mixed polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9FAA
 
 | | Cryo-EM structure of cardiac collagen-associated amyloid AL59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Helical superstructures between amyloid and collagen in cardiac fibrils from a patient with AL amyloidosis.
Nat Commun, 15, 2024
|
|
9FAB
 
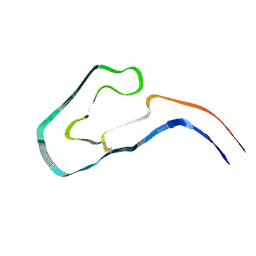 | | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal immunoglobulin light chains (LC) | | Authors: | Schulte, T, Speranzini, V, Chaves-Sanjuan, A, Milazzo, M, Ricagno, S. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Additional cryo-EM structure of cardiac amyloid AL59 - bent polymorph
To be published
|
|
7SVM
 
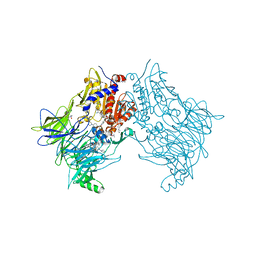 | | DPP8 IN COMPLEX WITH LIGAND ICeD-2 | | Descriptor: | (2S)-2-amino-1-(1,3-dihydro-2H-isoindol-2-yl)-2-[(1r,4S)-4-(pyrrolidin-1-yl)cyclohexyl]ethan-1-one, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVN
 
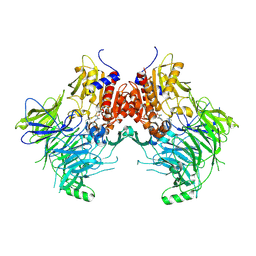 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
7SVO
 
 | | DPP8 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 8, trimethylamine oxide | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
