1YOI
 
 | | COBALT MYOGLOBIN (OXY) | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Brucker, E.A, Phillips Jr, G.N. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High resolution crystal structures of the deoxy, oxy, and aquomet forms of cobalt myoglobin.
J.Biol.Chem., 271, 1996
|
|
1YOG
 
 | | COBALT MYOGLOBIN (DEOXY) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING CO, SULFATE ION | | Authors: | Brucker, E.A, Phillips Jr, G.N. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High resolution crystal structures of the deoxy, oxy, and aquomet forms of cobalt myoglobin.
J.Biol.Chem., 271, 1996
|
|
1MNI
 
 | |
1MOC
 
 | |
1MOD
 
 | |
1MOA
 
 | |
1MOB
 
 | |
2RGF
 
 | | RBD OF RAL GUANOSINE-NUCLEOTIDE EXCHANGE FACTOR (PROTEIN), NMR, 10 STRUCTURES | | Descriptor: | RALGEF-RBD | | Authors: | Geyer, M, Herrmann, C, Wittinghofer, A, Kalbitzer, H.R. | | Deposit date: | 1997-02-13 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling.
Nat.Struct.Biol., 4, 1997
|
|
6C4D
 
 | | Structure based design of RIP1 kinase inhibitors | | Descriptor: | (3S)-3-(2-benzyl-3-chloro-7-oxo-2,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridin-6-yl)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepine-8-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
1N27
 
 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
4ZDK
 
 | | Crystal structure of the M. tuberculosis CTP synthase PyrG in complex with UTP, AMP-PCP and oxonorleucine | | Descriptor: | 5-OXO-L-NORLEUCINE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Bellinzoni, M, Barilone, N, Alzari, P.M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG.
Chem.Biol., 22, 2015
|
|
4ZDJ
 
 | | Crystal structure of the M. tuberculosis CTP synthase PyrG in complex with two UTP molecules | | Descriptor: | CTP synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bellinzoni, M, Barilone, N, Alzari, P.M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG.
Chem.Biol., 22, 2015
|
|
4ZDI
 
 | |
4JRF
 
 | |
6C3E
 
 | | CRYSTAL STRUCTURE OF RIP1 KINASE BOUND TO INHIBITOR | | Descriptor: | 2-benzyl-5-nitro-1H-benzimidazole, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Saikatendu, K.S, Yoshikawa, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 7-Oxo-2,4,5,7-tetrahydro-6 H-pyrazolo[3,4- c]pyridine Derivatives as Potent, Orally Available, and Brain-Penetrating Receptor Interacting Protein 1 (RIP1) Kinase Inhibitors: Analysis of Structure-Kinetic Relationships.
J. Med. Chem., 61, 2018
|
|
4JG5
 
 | |
5Y41
 
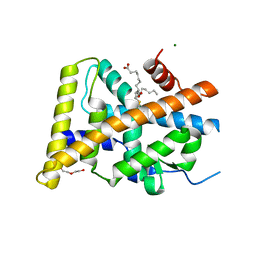 | | Crystal Structure of LIGAND-BOUND NURR1-LBD | | Descriptor: | (13E,15S)-15-hydroxy-9-oxoprosta-10,13-dien-1-oic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sreekanth, R, Lescar, J, Yoon, H.S. | | Deposit date: | 2017-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Nat.Chem.Biol., 2020
|
|
4K4K
 
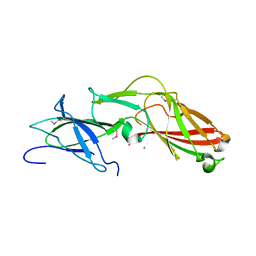 | |
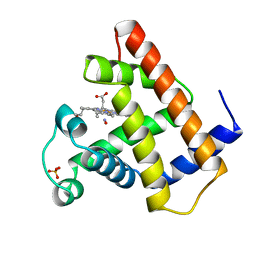
1HJT
 
 | |
3LIU
 
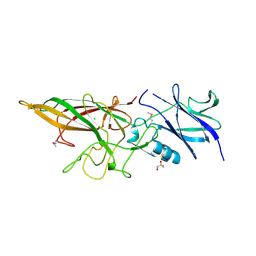 | |
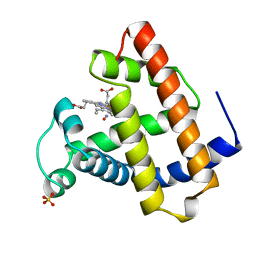
1JDO
 
 | |
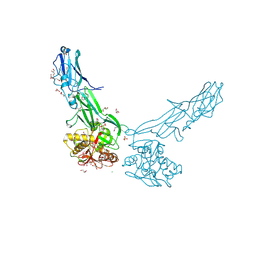
4EPS
 
 | |
7C3I
 
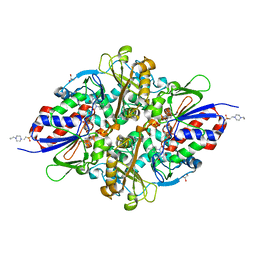 | | Structure of L-lysine oxidase D212A/D315A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7C3L
 
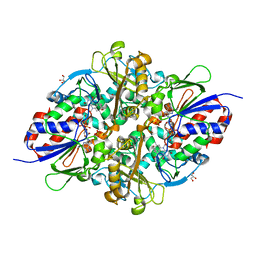 | | Structure of L-lysine oxidase D212A/D315A in complex with L-tyrosine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-lysine oxidase, ... | | Authors: | Kitagawa, M, Matsumoto, Y, Inagaki, K, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
7C3H
 
 | | Structure of L-lysine oxidase in complex with L-lysine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kondo, H, Kitagawa, M, Sugiyama, S, Imada, K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of strict substrate recognition of l-lysine alpha-oxidase from Trichoderma viride.
Protein Sci., 29, 2020
|
|
