7DDY
 
 | |
6M0W
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | Descriptor: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
1FBG
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBC
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBF
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
7E1T
 
 | | Crystal structure of Rab9A-GTP-Nde1 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoform 2 of Nuclear distribution protein nudE homolog 1, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Zhang, T, Ding, J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nde1 is a Rab9 effector for loading late endosomes to cytoplasmic dynein motor complex.
Structure, 30, 2022
|
|
2P6W
 
 | | Crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein | | Authors: | Zhang, Y, Ye, X, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
6M0V
 
 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-22 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0X
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | Descriptor: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | Authors: | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | Deposit date: | 2020-02-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
1FBD
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBE
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, ZINC ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBH
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
2P73
 
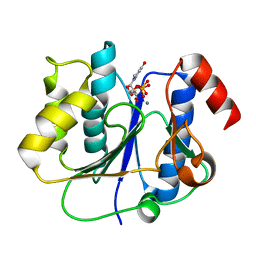 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
2P72
 
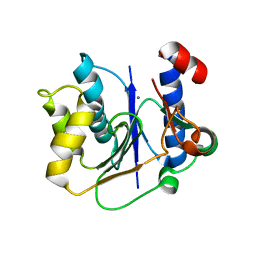 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
2MN6
 
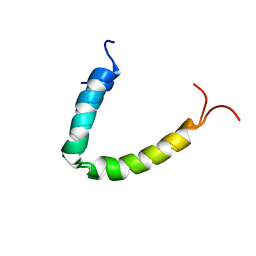 | |
7EU7
 
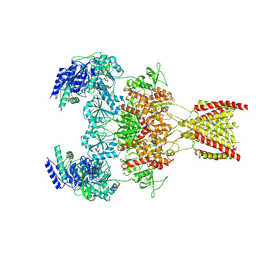 | | Structure of the human GluN1-GluN2A NMDA receptor in complex with S-ketamine, glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, Y, Zhang, T, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-04 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
6J1L
 
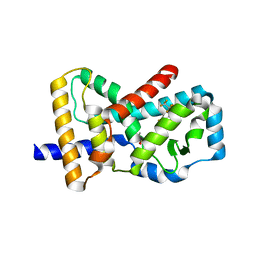 | | Crystal Structure Analysis of the ROR gamma(C455E) | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | Authors: | zhang, Y, Li, C.C, wu, X.S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
4OIP
 
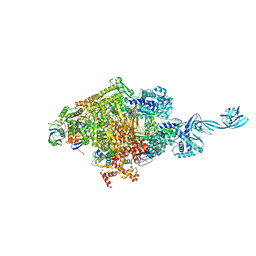 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIR
 
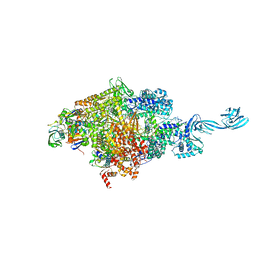 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4L1I
 
 | |
4L12
 
 | |
4L13
 
 | |
7E7S
 
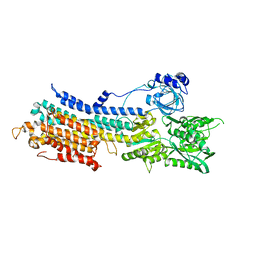 | | WT transporter state1 | | Descriptor: | CALCIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-02-27 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Embo J., 40, 2021
|
|
7D6E
 
 | |
