6DDL
 
 | | Crystal structure of the single mutant (D52N) of NT5C2-Q523X in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DE2
 
 | | Crystal structure of the double mutant (D52N/L375F) of the full-length NT5C2 in the active state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDY
 
 | | Crystal structure of the double mutant (D52N/K359Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDB
 
 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the basal state, Northeast Structural Genomics Consortium Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
3EVX
 
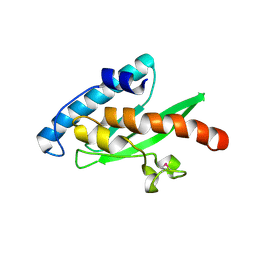 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
3LMO
 
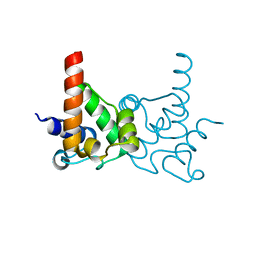 | | Crystal Structure of specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Forouhar, F, Rossi, P, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
5TI9
 
 | | Crystal structure of human TDO in complex with Trp and dioxygen, Northeast Structural Genomics Consortium Target HR6161 | | Descriptor: | N'-Formylkynurenine, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Forouhar, F, Lewis-Ballester, A, Lew, S, Karkashon, S, Seetharaman, J, Yeh, S.R, Tong, L. | | Deposit date: | 2016-10-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for catalysis and substrate-mediated cellular stabilization of human tryptophan 2,3-dioxygenase.
Sci Rep, 6, 2016
|
|
5TIA
 
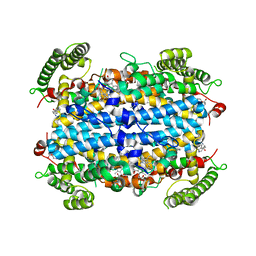 | | Crystal structure of human TDO in complex with Trp, Northeast Structural Genomics Consortium Target HR6161 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Lewis-Ballester, A, Lew, S, Karkashon, S, Seetharaman, J, Yeh, S.R, Tong, L. | | Deposit date: | 2016-10-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | Molecular basis for catalysis and substrate-mediated cellular stabilization of human tryptophan 2,3-dioxygenase.
Sci Rep, 6, 2016
|
|
2QGQ
 
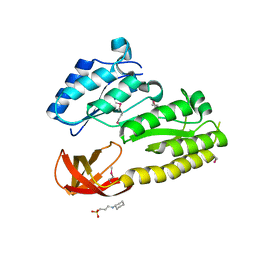 | | Crystal structure of TM_1862 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR77 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Protein TM_1862 | | Authors: | Forouhar, F, Neely, H, Hussain, M, Seetharaman, J, Fang, Y, Chen, C.X, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Post-translational Modification of Ribosomal Proteins: STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF RimO FROM THERMOTOGA MARITIMA, A RADICAL S-ADENOSYLMETHIONINE METHYLTHIOTRANSFERASE.
J.Biol.Chem., 285, 2010
|
|
2EVE
 
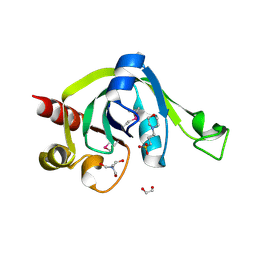 | | X-Ray Crystal Structure of Protein PSPTO5229 from Pseudomonas syringae. Northeast Structural Genomics Consortium Target PsR62 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Forouhar, F, Zhou, W, Belachew, A, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
1ZCE
 
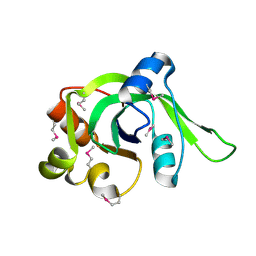 | | X-Ray Crystal Structure of Protein Atu2648 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR33. | | Descriptor: | hypothetical protein Atu2648 | | Authors: | Forouhar, F, Chen, Y, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
6MRO
 
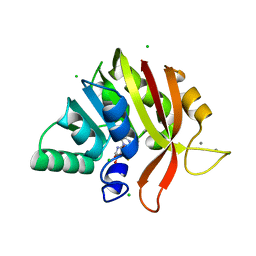 | | Crystal structure of methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution, Northeast Structural Genomics Consortium (NESG) Target MvR53. | | Descriptor: | CALCIUM ION, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Forouhar, F, Wang, C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-10-15 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution.
To Be Published
|
|
1UD0
 
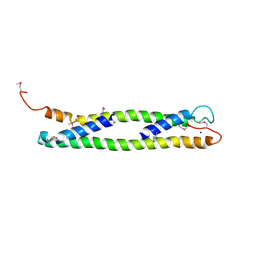 | | CRYSTAL STRUCTURE OF THE C-TERMINAL 10-kDA SUBDOMAIN OF HSC70 | | Descriptor: | 70 kDa heat-shock-like protein, SODIUM ION | | Authors: | Chou, C.C, Forouhar, F, Yeh, Y.H, Wang, C, Hsiao, C.D. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the C-terminal 10-kDa subdomain of Hsc70
J.BIOL.CHEM., 278, 2003
|
|
2IBO
 
 | | X-ray Crystal Structure of Protein SP2199 from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR31 | | Descriptor: | Hypothetical protein SP2199 | | Authors: | Seetharaman, J, Abashidze, M, Forouhar, F, Shastry, R, Conover, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein SP2199 from Streptococcus pneumoniae, Northeast structural genomics target SpR31
To be Published
|
|
2I2L
 
 | | X-ray Crystal Structure of Protein yopX from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR411. | | Descriptor: | YopX protein | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Forouhar, F, Kuzin, A.A, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein yopX from Bacillus subtilis
To be Published
|
|
2IMJ
 
 | | X-ray Crystal Structure of Protein PFL_3262 from Pseudomonas fluorescens. Northeast Structural Genomics Consortium Target PlR14. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Hypothetical protein DUF1348 | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the hypothetical protein (DUF1348) from Pseudomonas fluorescens, Northeast Structural Genomics target PlR14.
TO BE PUBLISHED
|
|
2IN5
 
 | | Crystal Structure of the hypothetical lipoprotein YmcC from Escherichia coli (K12), Northeast Structural Genomics target ER552. | | Descriptor: | Hypothetical lipoprotein ymcC | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Chen, C.X, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-05 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the hypothetical lipoprotein YmcC from Escherichia coli (K12), Northeast Structural Genomics target ER552.
TO BE PUBLISHED
|
|
1H65
 
 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
4OHF
 
 | | Crystal structure of cytosolic nucleotidase II (LPG0095) in complex with GMP from Legionella pneumophila, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET LGR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Srinivisan, B, Forouhar, F, Shukla, A, Sampangi, C, Kulkarni, S, Abashidze, M, Seetharaman, J, Lew, S, Mao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.M, Tong, L, Balaram, H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
3U1O
 
 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
7DMG
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | Descriptor: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | Descriptor: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLD
 
 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | Descriptor: | Carbonyl Reductase, MAGNESIUM ION | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DN1
 
 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
