3CFO
 
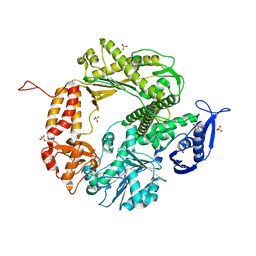 | | Triple Mutant APO structure | | Descriptor: | DNA polymerase, GUANOSINE, SULFATE ION | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
1KLN
 
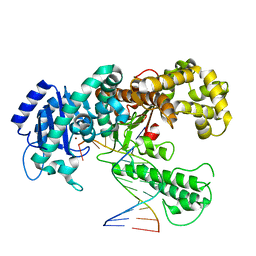 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*CP*GP*CP*GP*GP*CP*GP*GP*C)-3'), PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Beese, L.S, Derbyshire, V, Steitz, T.A. | | Deposit date: | 1994-05-24 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of DNA polymerase I Klenow fragment bound to duplex DNA.
Science, 260, 1993
|
|
1N8R
 
 | | Structure of large ribosomal subunit in complex with virginiamycin M | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10e, 50S ribosomal protein L13P, ... | | Authors: | Hansen, J.L, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-11-21 | | Release date: | 2003-07-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1NJI
 
 | | Structure of chloramphenicol bound to the 50S ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10e, 50S ribosomal protein L13P, ... | | Authors: | Hansen, J.L, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-12-31 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
4Z8C
 
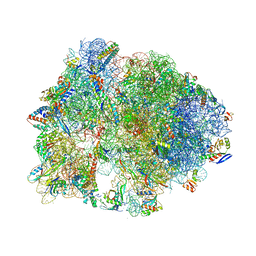 | | Crystal structure of the Thermus thermophilus 70S ribosome bound to translation inhibitor oncocin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Roy, R.N, Lomakin, I.B, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2015-04-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4Z3S
 
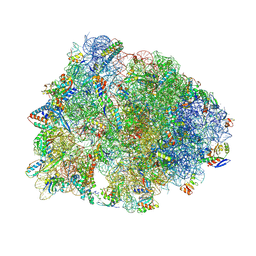 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic A201A, mRNA and three tRNAs in the A, P and E sites at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 3'-deoxy-3'-{[(2E)-3-(4-{[(4Z)-6-O-(6-deoxy-3,4-di-O-methyl-alpha-D-mannopyranosyl)-5-O-methyl-alpha-D-threo-hex-4-enofuranosyl]oxy}phenyl)-2-methylprop-2-enoyl]amino}-N,N-dimethyladenosine, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-03-31 | | Release date: | 2015-06-03 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
1D89
 
 | |
1IG9
 
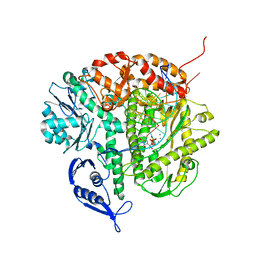 | | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*(DOC))-3', CALCIUM ION, ... | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-06-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
1GSG
 
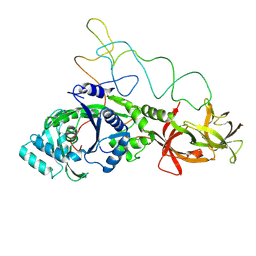 | | Structure of E.coli glutaminyl-tRNA synthetase complexed with trnagln and ATP at 2.8 Angstroms resolution | | Descriptor: | GLUTAMINYL-TRNA SYNTHETASE, TRNAGLN | | Authors: | Rould, M.A, Perona, J.J, Soell, D, Steitz, T.A. | | Deposit date: | 1990-04-03 | | Release date: | 1992-02-24 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution.
Science, 246, 1989
|
|
1IH7
 
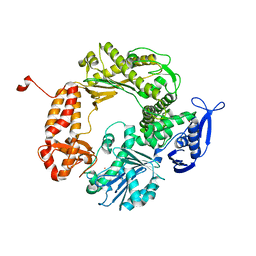 | | High-Resolution Structure of Apo RB69 DNA Polymerase | | Descriptor: | DNA POLYMERASE, GUANOSINE, POTASSIUM ION | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Replicating Complex of a Pol alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
1K73
 
 | | Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, ANISOMYCIN, ... | | Authors: | Hansen, J, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1G6N
 
 | | 2.1 ANGSTROM STRUCTURE OF CAP-CAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CATABOLITE GENE ACTIVATOR PROTEIN | | Authors: | Passner, J.M, Schultz, S.C, Steitz, T.A. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution.
J.Mol.Biol., 304, 2000
|
|
354D
 
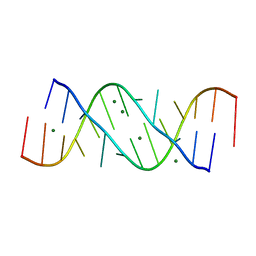 | | Structure of loop E FROM E. coli 5S RRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*G)-3'), RNA-DNA (5'-R(*GP*CP*GP*AP*GP*AP*GP*UP*AP*)-D(*DGP(S)*)-R(*GP*C)-3') | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-10-01 | | Release date: | 1997-11-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
357D
 
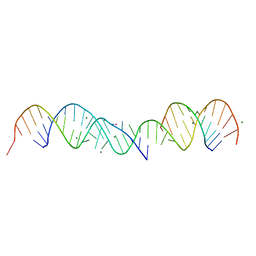 | | 3.5 A structure of fragment I from E. coli 5S RRNA | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, RNA (5'-R(*CP*CP*CP*CP*AP*UP*GP*CP*GP*AP*GP*AP*GP*UP*AP*GP*G P*GP*AP*AP*CP*UP* GP*CP*CP*AP*GP*GP*CP*AP*U)-3'), ... | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-10-09 | | Release date: | 1997-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
364D
 
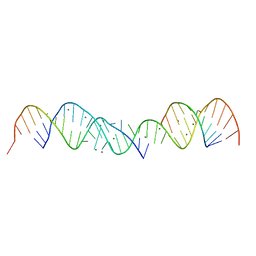 | | 3.0 A STRUCTURE OF FRAGMENT I FROM E. COLI 5S RRNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*CP*CP*CP*AP*UP*GP*CP*GP*AP*GP*AP*GP*UP*AP*GP*G P*GP*AP*AP*CP*UP*GP*CP*CP*AP*GP*GP*CP*AP*U)-3'), RNA (5'-R(*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*GP*UP*GP*GP*GP*G *UP*C)-3'), ... | | Authors: | Correll, C.C, Freeborn, B, Moore, P.B, Steitz, T.A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain.
Cell(Cambridge,Mass.), 91, 1997
|
|
1GTS
 
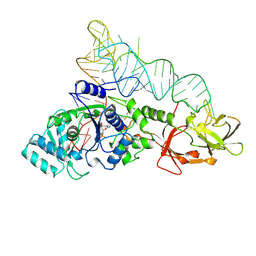 | |
1JJ2
 
 | | Fully Refined Crystal Structure of the Haloarcula marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The kink-turn: a new RNA secondary structure motif.
EMBO J., 20, 2001
|
|
1HVU
 
 | |
1KFS
 
 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*AP*CP*G)-3'), MAGNESIUM ION, PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1CGP
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*GP*TP*GP*TP*GP*AP*CP*AP*TP*AP*T)-3'), DNA (5'-D(*GP*TP*CP*AP*CP*AP*CP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Schultz, S.C, Shields, G.C, Steitz, T.A. | | Deposit date: | 1991-08-12 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees.
Science, 253, 1991
|
|
1EFG
 
 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1994-10-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
1CSL
 
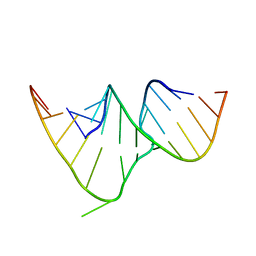 | | CRYSTAL STRUCTURE OF THE RRE HIGH AFFINITY SITE | | Descriptor: | 5'-R(*AP*AP*CP*GP*GP*GP*CP*GP*CP*AP*GP*AP*A)-3', 5'-R(*UP*CP*UP*GP*AP*CP*GP*GP*UP*AP*CP*GP*UP*UP*U)-3' | | Authors: | Ippolito, J.A, Steitz, T.A. | | Deposit date: | 1999-08-18 | | Release date: | 2000-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the HIV-1 RRE high affinity rev binding site at 1.6 A resolution.
J.Mol.Biol., 295, 2000
|
|
1CEZ
 
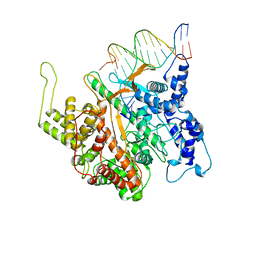 | | CRYSTAL STRUCTURE OF A T7 RNA POLYMERASE-T7 PROMOTER COMPLEX | | Descriptor: | DNA (5'-D(P*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3'), PROTEIN (BACTERIOPHAGE T7 RNA POLYMERASE) | | Authors: | Cheetham, G.M.T, Jeruzalmi, D, Steitz, T.A. | | Deposit date: | 1999-03-11 | | Release date: | 1999-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for initiation of transcription from an RNA polymerase-promoter complex.
Nature, 399, 1999
|
|
1FFY
 
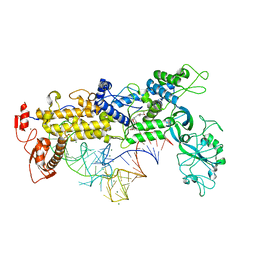 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 2000-07-26 | | Release date: | 2000-08-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
1CLQ
 
 | |
