1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
2G97
 
 | |
2G96
 
 | |
6DM8
 
 | | Understanding the Species Selectivity of Myeloid cell leukemia-1 (Mcl-1) inhibitors | | Descriptor: | 4-{8-chloro-11-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1-oxo-7-(1,3,5-trimethyl-1H-pyrazol-4-yl)-4,5-dihydro-1H-[1,4]diazepino[1,2-a]indol-2(3H)-yl}-1-methyl-1H-indole-6-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog - MBP chimera, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Understanding the Species Selectivity of Myeloid Cell Leukemia-1 (Mcl-1) Inhibitors.
Biochemistry, 57, 2018
|
|
7BYO
 
 | | Lysozyme structure SS1 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7BYP
 
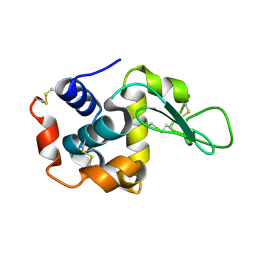 | | Lysozyme structure SASE1 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
4FQR
 
 | |
4FNL
 
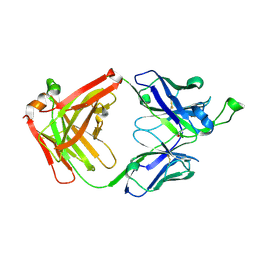 | |
4FQM
 
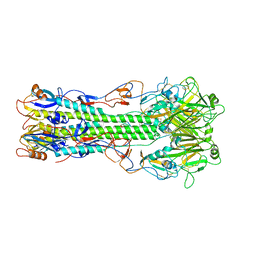 | | Structure of B/Brisbane/60/2008 Influenza Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQL
 
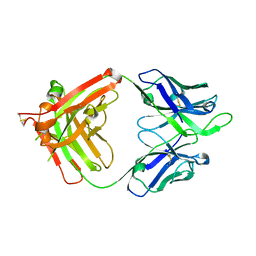 | | Influenza B HA Antibody (Fab) CR8033 | | Descriptor: | monoclonal antibody CR8033 heavy chain, monoclonal antibody CR8033 light chain | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQV
 
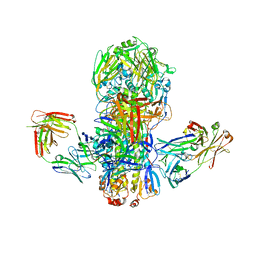 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQH
 
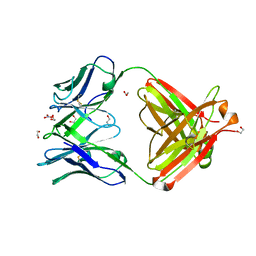 | | Crystal Structure of Fab CR9114 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, antibody CR9114 heavy chain, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FNK
 
 | |
5CUF
 
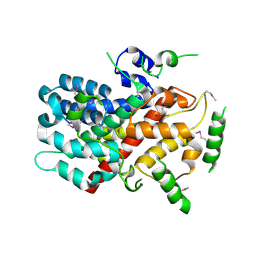 | | X-ray crystal structure of SeMet human Sestrin2 | | Descriptor: | Sestrin-2 | | Authors: | Kim, H, An, S, Ro, S.-H, Lee, J.H, Cho, U.-S. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains.
Nat Commun, 6, 2015
|
|
5K53
 
 | |
5D6Y
 
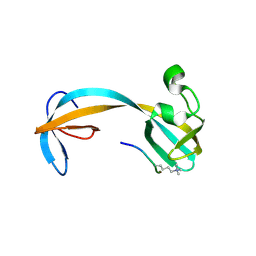 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5D6W
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5D6X
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
7BXZ
 
 | |
4UC4
 
 | |
4IVI
 
 | | Crystal structure of a family VIII carboxylesterase. | | Descriptor: | Carboxylesterase, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
4IVK
 
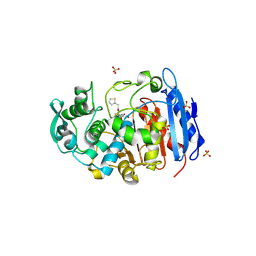 | | Crystal structure of a fammily VIII carboxylesterase in a complex with cephalothin. | | Descriptor: | CEPHALOTHIN GROUP, Carboxylesterases, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
6LKS
 
 | | Effects of zinc ion on oligomerization and pH stability of influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Seok, J, Kim, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Divalent cation-induced conformational changes of influenza virus hemagglutinin.
Sci Rep, 10, 2020
|
|
6FRK
 
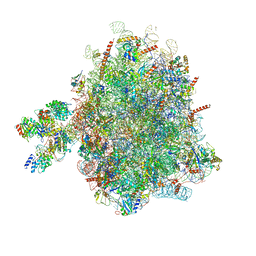 | | Structure of a prehandover mammalian ribosomal SRP and SRP receptor targeting complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kobayashi, K, Jomaa, A, Ban, N. | | Deposit date: | 2018-02-16 | | Release date: | 2018-03-28 | | Last modified: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex.
Science, 360, 2018
|
|
8HNP
 
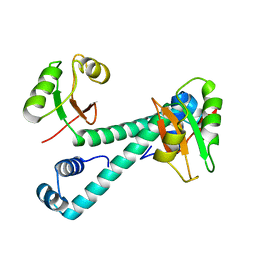 | | Archaeal transcription factor Mutant | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
