6KR6
 
 | | Crystal structure of Drosophila Piwi | | Descriptor: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | Authors: | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
8WUP
 
 | |
8WW5
 
 | |
8WWT
 
 | |
8WX6
 
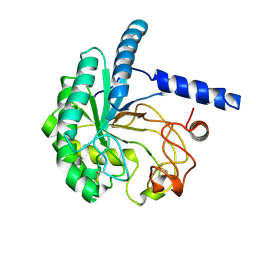 | |
7V6C
 
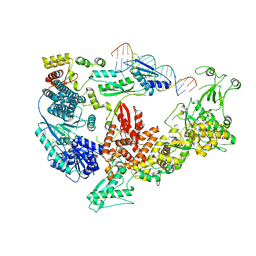 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6B
 
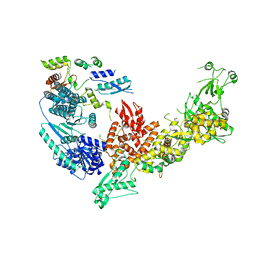 | | Structure of the Dicer-2-R2D2 heterodimer | | Descriptor: | Dicer-2, isoform A, R2D2 | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
2ZOI
 
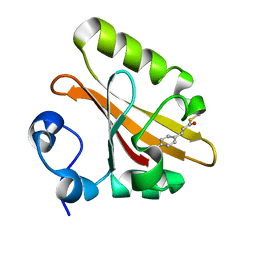 | |
2ZOH
 
 | |
1TIA
 
 | | AN UNUSUAL BURIED POLAR CLUSTER IN A FAMILY OF FUNGAL LIPASES | | Descriptor: | LIPASE | | Authors: | Derewenda, U, Swenson, L, Yamaguchi, S, Wei, Y, Derewenda, Z.S. | | Deposit date: | 1993-12-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unusual buried polar cluster in a family of fungal lipases.
Nat.Struct.Biol., 1, 1994
|
|
3ABG
 
 | | X-ray Crystal Analysis of Bilirubin Oxidase from Myrothecium verrucaria at 2.3 angstrom Resolution using a Twin Crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Mizutani, K, Toyoda, M, Sagara, K, Takahashi, N, Sato, A, Kamitaka, Y, Tsujimura, S, Nakanishi, Y, Sugiura, T, Yamaguchi, S, Kano, K, Mikami, B. | | Deposit date: | 2009-12-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3W05
 
 | | Crystal structure of Oryza sativa DWARF14 (D14) in complex with PMSF | | Descriptor: | 1,2-ETHANEDIOL, Dwarf 88 esterase, phenylmethanesulfonic acid | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W06
 
 | | Crystal structure of Arabidopsis thaliana DWARF14 Like (AtD14L) | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase, alpha/beta fold family protein | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W04
 
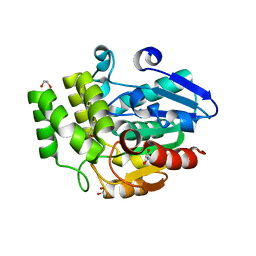 | | Crystal structure of Oryza sativa DWARF14 (D14) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3A56
 
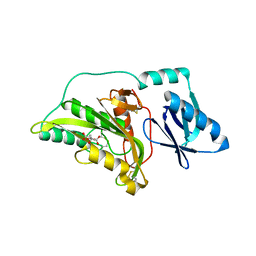 | |
3A55
 
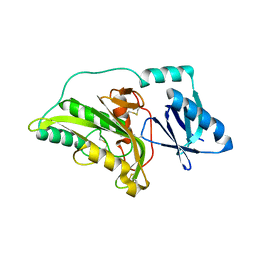 | |
3A54
 
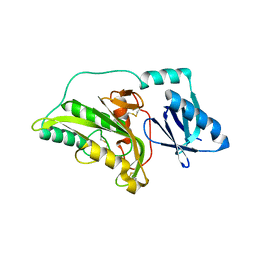 | |
3A73
 
 | |
2Y3W
 
 | |
8HD2
 
 | |
1TIB
 
 | |
1TIC
 
 | | CONFORMATIONAL LABILITY OF LIPASES OBSERVED IN THE ABSENCE OF AN OIL-WATER INTERFACE: CRYSTALLOGRAPHIC STUDIES OF ENZYMES FROM THE FUNGI HUMICOLA LANUGINOSA AND RHIZOPUS DELEMAR | | Descriptor: | LIPASE | | Authors: | Derewenda, U, Swenson, L, Green, R, Joerger, R, Haas, M.J, Derewenda, Z.S. | | Deposit date: | 1993-12-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational lability of lipases observed in the absence of an oil-water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar.
J.Lipid Res., 35, 1994
|
|
2Y3V
 
 | | N-terminal head domain of Danio rerio SAS-6 | | Descriptor: | SPINDLE ASSEMBLY ABNORMAL PROTEIN 6 HOMOLOG | | Authors: | van Breugel, M. | | Deposit date: | 2010-12-27 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of SAS-6 suggest its organization in centrioles.
Science, 331, 2011
|
|
4YPJ
 
 | | X-ray Structure of The Mutant of Glycoside Hydrolase | | Descriptor: | Beta galactosidase | | Authors: | Ishikawa, K, Kataoka, M, Yanamoto, T, Nakabayashi, M, Watanabe, M. | | Deposit date: | 2015-03-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of beta-galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants.
Febs J., 282, 2015
|
|
6AZB
 
 | |
