8AJU
 
 | | Crystal structure of the Q65A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Crystal structure of the Q65A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AJV
 
 | | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa crystallized in the presence of K+ cations | | Descriptor: | Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa crystallized in the presence of K+ cations
To Be Published
|
|
8AJT
 
 | | Crystal structure of the H323A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of the H323A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AJW
 
 | | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AZF
 
 | | Crystal structure of K449E variant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa in complex with adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Arning, A, Malecki, P, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of K449E variant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa in complex with adenosine
To Be Published
|
|
7ZD8
 
 | | Crystal structure of the R24E mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD7
 
 | | Crystal structure of the R24E/E352T double mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD9
 
 | | Crystal structure of the E352T mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8AJS
 
 | | Crystal structure of the F324A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of the F324A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
5HLM
 
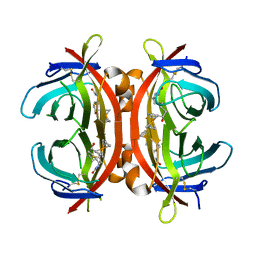 | | Crystal structure of avidin complex with a ferrocene biotin derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin, [(1,2,3,4,5-eta)-cyclopentadienyl]{(1,2,3,4,5-eta)-1-[1-hydroxy-5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentyl]cyclopentadienyl}iron | | Authors: | Strzelczyk, P, Bujacz, G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ferrocene-Biotin Conjugates: Synthesis, Structure, Cytotoxic Activity and Interaction with Avidin
Chempluschem, 81, 2016
|
|
5J23
 
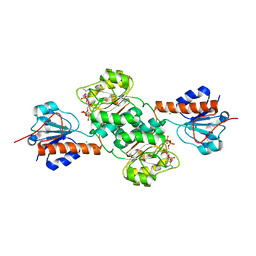 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with 2'-phospho-ADP-ribose | | Descriptor: | 2-hydroxyacid dehydrogenase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Kutner, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
7O5M
 
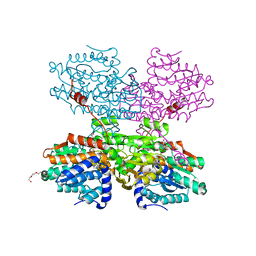 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Na+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7O5L
 
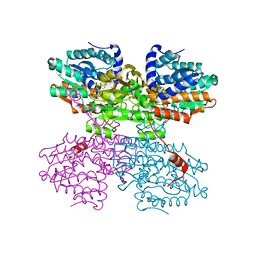 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4WEQ
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADP and sulfate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Osinski, T, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
4Z0P
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Porebski, P.J, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5ELY
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU242 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{R})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-11-05 | | Release date: | 2016-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
6BII
 
 | | Crystal Structure of Pyrococcus yayanosii Glyoxylate Hydroxypyruvate Reductase in complex with NADP and malonate (re-refinement of 5AOW) | | Descriptor: | GLYCEROL, Glyoxylate reductase, MALONATE ION, ... | | Authors: | Lassalle, L, Shabalin, I.G, Girard, E, Minor, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases.
Sci Rep, 6, 2016
|
|
7QGC
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 5.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CITRATE ANION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGE
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6,7,8-TETRABROMOBENZOTRIAZOLE (TBBt) AT PH 8.5 | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGD
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 8.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
7QGB
 
 | | H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 6.5 | | Descriptor: | 5,6-DIBROMOBENZOTRIAZOLE, Casein kinase II subunit alpha | | Authors: | Winiewska-Szajewska, M, Czapinska, H, Kaus-Drobek, M, Piasecka, A, Mieczkowska, K, Dadlez, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Competition between electrostatic interactions and halogen bonding in the protein-ligand system: structural and thermodynamic studies of 5,6-dibromobenzotriazole-hCK2 alpha complexes.
Sci Rep, 12, 2022
|
|
5UNN
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH-dependent glyoxylate/hydroxypyruvate reductase | | Authors: | Shabalin, I.G, LaRowe, C, Kutner, J, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V6Q
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and malonate | | Descriptor: | MALONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent glyoxylate/hydroxypyruvate reductase, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miezaniec, A.P, Gasiorowska, O.A, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V72
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with citrate | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5UOG
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in apo form | | Descriptor: | NADPH-dependent glyoxylate/hydroxypyruvate reductase, SULFATE ION | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
