6ASM
 
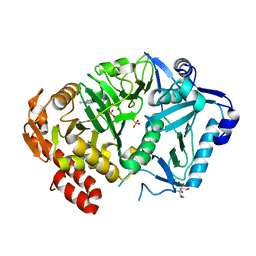 | | E. coli phosphoenolpyruvate carboxykinase G209S K212C mutant bound to thiosulfate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT4
 
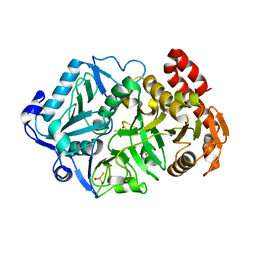 | |
6AT3
 
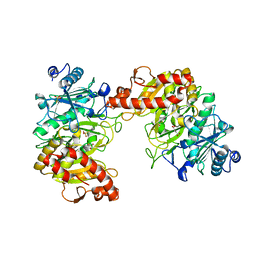 | |
6ASI
 
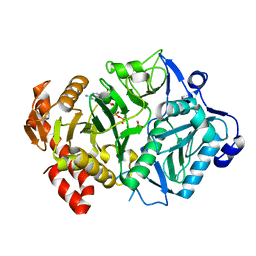 | | E. coli phosphoenolpyruvate carboxykinase G209S mutant bound to methanesulfonate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT2
 
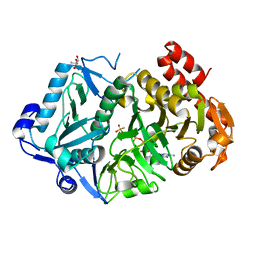 | | E. coli phosphoenolpyruvate carboxykinase G209N mutant bound to thiosulfate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | Deposit date: | 2017-08-27 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6ASN
 
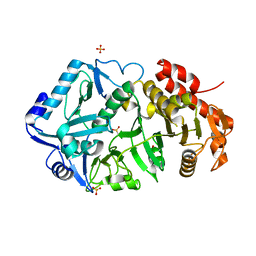 | |
3J7A
 
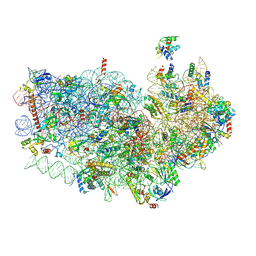 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, small subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
3J79
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
3JBO
 
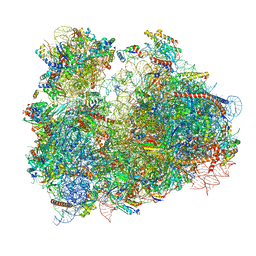 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P/E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBP
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JBN
 
 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to P-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
6OKK
 
 | |
5V4M
 
 | | Structure of HLA-DR15 with bound alpha3(135-145) peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA-DRA1, ... | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells.
Nature, 545, 2017
|
|
5V4N
 
 | | Structure of HLA-DR1 with bound alpha3(135-145) peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA-DRA1, alpha3(135-145)-HLA-DRB1*01:01 | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2017-03-10 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells.
Nature, 545, 2017
|
|
2RH4
 
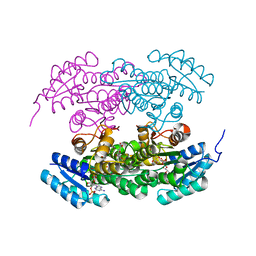 | | Actinorhodin ketoreductase, actKR, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2RHC
 
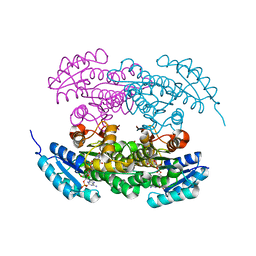 | | Actinorhodin ketordeuctase, actKR, with NADP+ and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-08 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2RHR
 
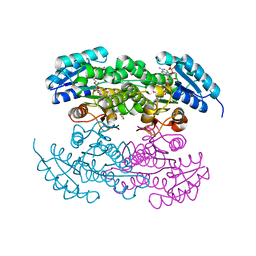 | | P94L actinorhodin ketordeuctase mutant, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-09 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2PCX
 
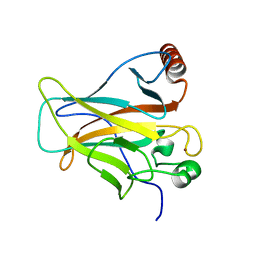 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1TGR
 
 | | Crystal Structure of mini-IGF-1(2) | | Descriptor: | Insulin-like growth factor IA | | Authors: | Liang, D.C, Yun, C.H, Chang, W.R. | | Deposit date: | 2004-05-29 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42A crystal structure of mini-IGF-1(2): an analysis of the disulfide isomerization property and receptor binding property of IGF-1 based on the three-dimensional structure
Biochem.Biophys.Res.Commun., 326, 2004
|
|
