7OLE
 
 | | Cryo-EM structure of the TELO2-TTI1-TTI2-RUVBL1-RUVBL2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2, ... | | Authors: | Pal, M, Llorca, O, Pearl, L. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-07 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone.
Cell Rep, 36, 2021
|
|
6FM8
 
 | | RPAP3 c-terminus | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Pal, M, Roe, S.M. | | Deposit date: | 2018-01-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex
Nat Commun, 2020
|
|
3NFT
 
 | | Near-atomic resolution analysis of BipD- A component of the type-III secretion system of Burkholderia pseudomallei | | Descriptor: | Translocator protein bipD | | Authors: | Pal, M, Erskine, P.T, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Near-atomic resolution analysis of BipD, a component of the type III secretion system of Burkholderia pseudomallei.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6X24
 
 | | Structural basis of plant blue light photoreceptor | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Palayam, M, Ganapathy, J, Guercio, M.A, Shabek, N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into photoactivation of plant Cryptochrome-2.
Commun Biol, 4, 2021
|
|
5KKV
 
 | |
5HHD
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D-Peptide RFX037.D, D-Vascular endothelial growth factor, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uppalapati, M, LEE, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5HHC
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D- Vascular endothelial growth factor-A, GLYCEROL, Vascular endothelial growth factor A | | Authors: | Uppalapati, M, Lee, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5WVX
 
 | | Crystal Structure of bifunctional Kunitz type Trypsin /amylase inhibitor (AMTIN) from the tubers of Alocasia macrorrhiza | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CITRIC ACID, Trypsin/chymotrypsin inhibitor | | Authors: | Palayam, M, Radhakrishnan, M, Lakshminarayanan, K, Balu, K.E, Ganapathy, J, Krishnasamy, G. | | Deposit date: | 2016-12-29 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural insights into a multifunctional inhibitor, 'AMTIN' from tubers of Alocasia macrorrhizos and its possible role in dengue protease (NS2B-NS3) inhibition.
Int. J. Biol. Macromol., 113, 2018
|
|
7EH3
 
 | |
4CHH
 
 | | N-terminal domain of yeast PIH1p | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN INTERACTING WITH HSP90 1 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGU
 
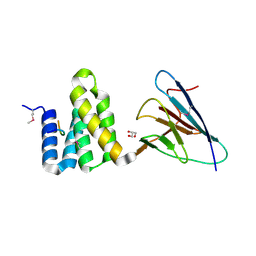 | | Full length Tah1 bound to yeast PIH1 and HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, PROTEIN INTERACTING WITH HSP90 1, ... | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGV
 
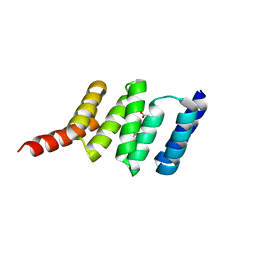 | | First TPR of Spaghetti (RPAP3) bound to HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, RNA POLYMERASE II-ASSOCIATED PROTEIN 3 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGW
 
 | |
7SA1
 
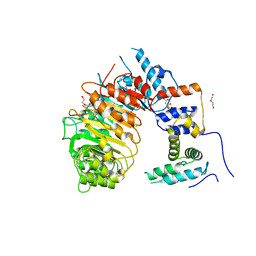 | | LRR-F-Box plant ubiquitin ligase | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, F-box/LRR-repeat MAX2 homolog, ... | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2021-09-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling.
Nat.Plants, 8, 2022
|
|
7UPV
 
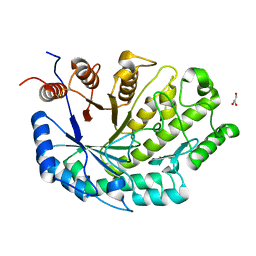 | |
4CGQ
 
 | | Full length Tah1 bound to HSP90 peptide SRMEEVD | | Descriptor: | CHLORIDE ION, HEAT SHOCK PROTEIN HSP 90-ALPHA, TPR REPEAT-CONTAINING PROTEIN ASSOCIATED WITH HSP90 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tah1 Helix-Swap Dimerization Prevents Mixed Hsp90 Co-Chaperone Complexes
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2W1P
 
 | | 1.4 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 8.0 | | Descriptor: | AQUAPORIN PIP2-7 7;, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-20 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism
Plos Biol., 7, 2009
|
|
2W2E
 
 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | Descriptor: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
7B00
 
 | | Human LAT2-4F2hc complex in the apo-state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Isoform 2 of 4F2 cell-surface antigen heavy chain, ... | | Authors: | Rodriguez, C.F, Escudero-Bravo, P, Garcia-Martin, C, Boskovic, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2020-11-17 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for substrate specificity of heteromeric transporters of neutral amino acids.
Proc Natl Acad Sci U S A, 118, 2021
|
|
8OFK
 
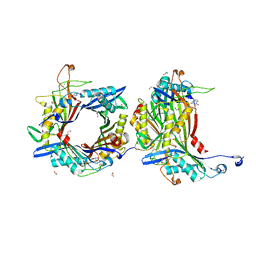 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under reducing conditions (space group C 2 2 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
8OIH
 
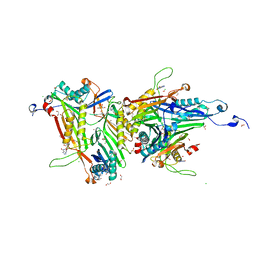 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under oxidising conditions (space group C 2 2 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, BROMIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
5BNG
 
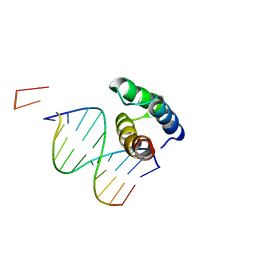 | | monomer of TALE type homeobox transcription factor MEIS1 complexes with specific DNA | | Descriptor: | DNA (5'-D(P*AP*AP*TP*TP*AP*GP*CP*TP*GP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(P*TP*GP*AP*CP*AP*GP*CP*TP*AP*A-3'), ... | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Nitta, K, Dave, K, Popov, A, Taipale, M, Enge, M, Kivioja, T, Taipale, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
8OH8
 
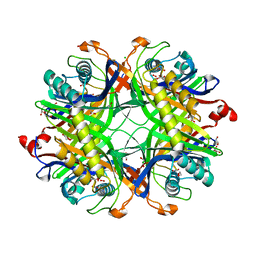 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under reducing conditions (space group P 21 21 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
8OIW
 
 | | Crystal structure of the cysteine-rich Gallus gallus urate oxidase in complex with the 8-azaxanthine inhibitor under oxidising conditions (space group P 21 21 21) | | Descriptor: | 1,2-ETHANEDIOL, 8-AZAXANTHINE, CHLORIDE ION, ... | | Authors: | Di Palma, M, Chegkazi, M, Bui, S, Mori, G, Percudani, R, Steiner, R.A. | | Deposit date: | 2023-03-23 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cysteine Enrichment Mediates Co-Option of Uricase in Reptilian Skin and Transition to Uricotelism.
Mol.Biol.Evol., 40, 2023
|
|
1NPJ
 
 | | Crystal structure of H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution of the type-1 active site of the H145G/A variants of nitrite reductase by ligand insertion
Biochemistry, 42, 2003
|
|
