7C95
 
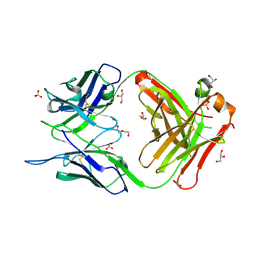 | | Crystal structure of the anti-human podoplanin antibody Fab fragment | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Nakamura, S, Suzuki, K, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7C94
 
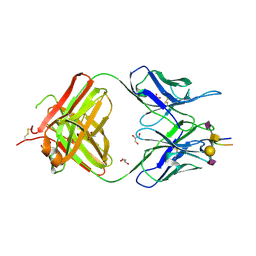 | | Crystal structure of the anti-human podoplanin antibody Fab fragment complex with glycopeptide | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | Authors: | Suzuki, K, Nakamura, S, Ogasawara, S, Naruchi, K, Shimabukuro, J, Tukahara, N, Kaneko, M.K, Kato, Y, Murata, T. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of an anti-podoplanin antibody bound to a disialylated O-linked glycopeptide.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYP
 
 | | Cryo-EM Structure of E-4031-bound hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2, ~{N}-[4-[1-[2-(6-methylpyridin-2-yl)ethyl]piperidin-4-yl]carbonylphenyl]methanesulfonamide | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYN
 
 | | Cryo-EM Structure of inhibitor-free hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYO
 
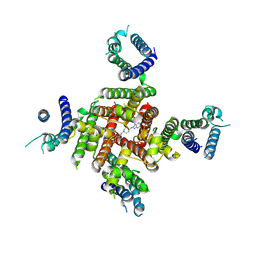 | | Cryo-EM Structure of astemizole-bound hERG Channel | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
5GIR
 
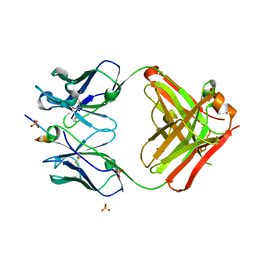 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of Fab fragment, LYS-PRO-ILE-ILE-ILE-GLY-SER-HIS-ALA-TYR-GLY-ASP, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GIS
 
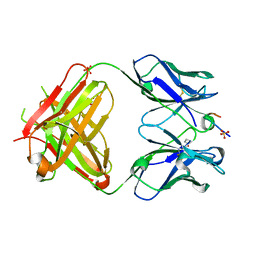 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, SULFATE ION, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
4CAD
 
 | | Mechanism of farnesylated CAAX protein processing by the integral membrane protease Rce1 | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kulkarni, K, Manolaridis, I, Dodd, R.B, Cronin, N, Ogasawara, S, Iwata, S, Barford, D. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Farnesylated Caax Protein Processing by the Intramembrane Protease Rce1
Nature, 504, 2013
|
|
7P34
 
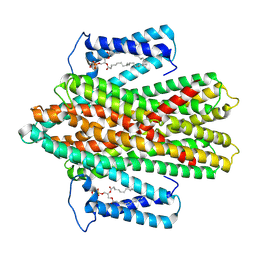 | |
4KPP
 
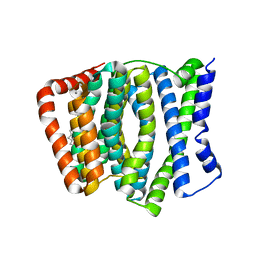 | | Crystal Structure of H+/Ca2+ Exchanger CAX | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, OLEIC ACID, ... | | Authors: | Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the counter-transport mechanism of a H+/Ca2+ exchanger.
Science, 341, 2013
|
|
7BTW
 
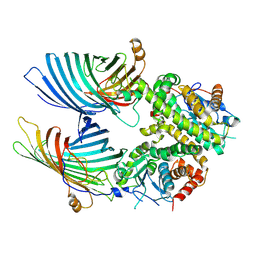 | | The mitochondrial SAM complex from S.cere | | Descriptor: | Mitochondrial outer membrane beta-barrel protein, SAM37 isoform 1, Sorting assembly machinery 35 kDa subunit | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTX
 
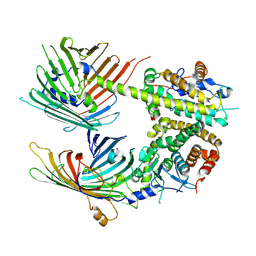 | | The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTY
 
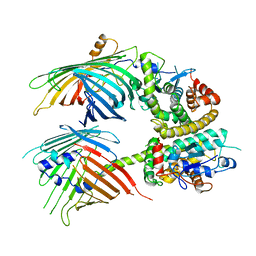 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
8WCI
 
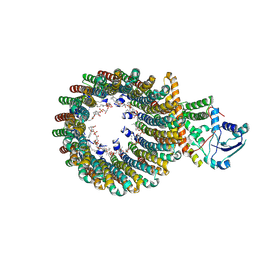 | | Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae | | Descriptor: | CARDIOLIPIN, N,N-dimethyl-4-(5-methyl-1H-benzimidazol-2-yl)aniline, SODIUM ION, ... | | Authors: | Suzuki, K, Mikuriya, S, Adachi, N, Kawasaki, M, Senda, T, Moriya, T, Murata, T. | | Deposit date: | 2023-09-12 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Novel Inhibitor of Na+-Transporting V-ATPase Suppresses VRE Colonization in Mice and Reveals the High-Resolution Structure of the Na+ Transport Pathway
To Be Published
|
|
7CN0
 
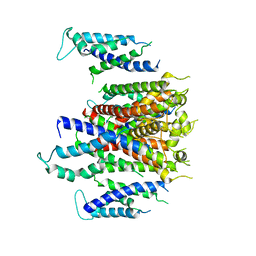 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CN1
 
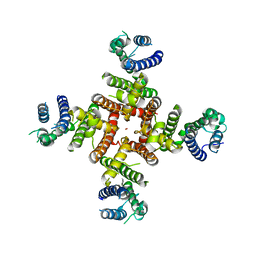 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | Descriptor: | POTASSIUM ION, potassium channel | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7VPK
 
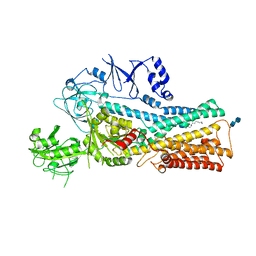 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPL
 
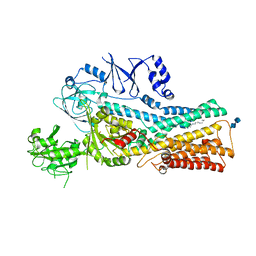 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2Pi state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPI
 
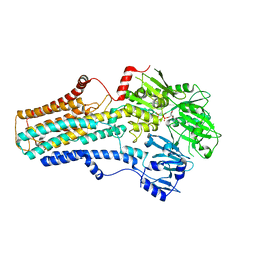 | | Cryo-EM structure of the human ATP13A2 (E1-ATP state) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Polyamine-transporting ATPase 13A2 | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPJ
 
 | | Cryo-EM structure of the human ATP13A2 (E1P-ADP state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
8Y6H
 
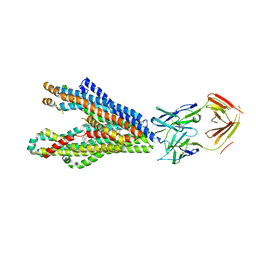 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
4YNY
 
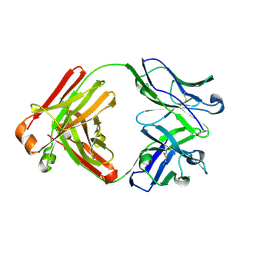 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 | | Descriptor: | Heavy chain of antigen binding fragment, Fab, Light chain of antigen binding fragment | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
4YO0
 
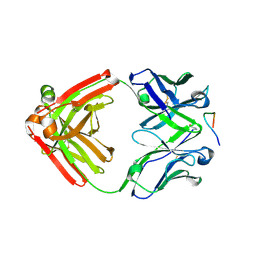 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 with bound PA peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antigen binding fragment, Fab, ... | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
