6LKD
 
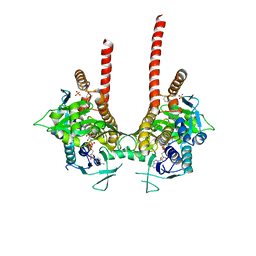 | | in meso full-length rat KMO in complex with a pyrazoyl benzoic acid inhibitor | | Descriptor: | 5-[5-(4-chloranyl-3-fluoranyl-phenyl)-4-methyl-pyrazol-1-yl]-2-phenylmethoxy-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
6LKE
 
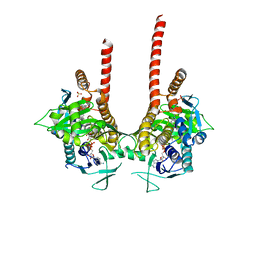 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
3ABU
 
 | | Crystal Structure of LSD1 in complex with a 2-PCPA derivative, S1201 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3R,3aS)-3-[2-(benzyloxy)-3-fluorophenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
2Z3Y
 
 | | Crystal structure of Lysine-specific demethylase1 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Mimasu, S, Sengoku, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-08 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
2Z5U
 
 | |
5Y9T
 
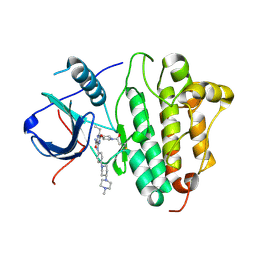 | | Crystal Structure of EGFR T790M mutant in complex with naquotinib | | Descriptor: | 6-ethyl-3-[[4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-[(3R)-1-prop-2-enoylpyrrolidin-3-yl]oxy-pyrazin e-2-carboxamide, Epidermal growth factor receptor | | Authors: | Mimasu, S, Tomimoto, Y, Maiko, I, Yasushi, A, Tatsuya, N. | | Deposit date: | 2017-08-28 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, inEGFR-Mutated Non-Small Cell Lung Cancer.
Mol. Cancer Ther., 17, 2018
|
|
2EJR
 
 | | LSD1-tranylcypromine complex | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Sengoku, T, Mimasu, S, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
3AJI
 
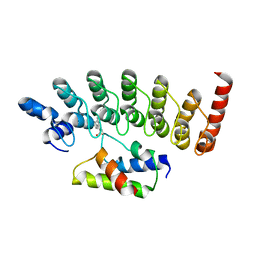 | | Structure of Gankyrin-S6ATPase photo-cross-linked site-specifically, and incoporated by genetic code expansion | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, Proteasome (Prosome, macropain) 26S subunit, ... | | Authors: | Sato, S, Mimasu, S, Sato, A, Hino, N, Sakamoto, K, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-07 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid
Biochemistry, 50, 2011
|
|
2DW4
 
 | |
