1K4W
 
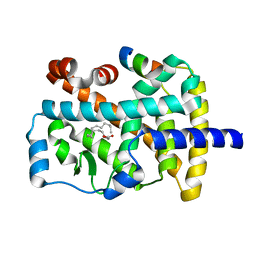 | | X-ray structure of the orphan nuclear receptor ROR beta ligand-binding domain in the active conformation | | Descriptor: | Nuclear receptor ROR-beta, STEARIC ACID, steroid receptor coactivator-1 | | Authors: | Stehlin, C, Wurtz, J.M, Steinmetz, A, Greiner, E, Schuele, R, Moras, D, Renaud, J.P. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation.
EMBO J., 20, 2001
|
|
8HUS
 
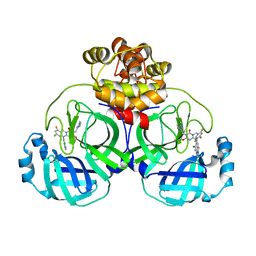 | | Crystal structure of SARS main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUT
 
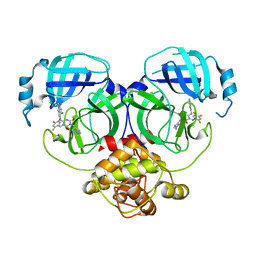 | | Crystal structure of MERS main protease in complex with S217622 | | Descriptor: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8IG5
 
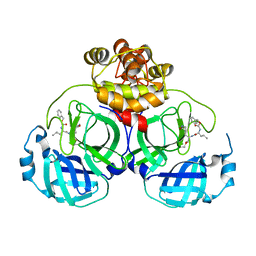 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
6CCW
 
 | |
8WVW
 
 | | Cryo-EM structure of LGR4 in state II | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4 | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of LGR4 in state I
To Be Published
|
|
8WVY
 
 | |
8WVV
 
 | | Cryo-EM structure of LGR4 in state I | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4 | | Authors: | Lin, C, Chang, Z. | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of LGR4 in state I
To Be Published
|
|
7WQI
 
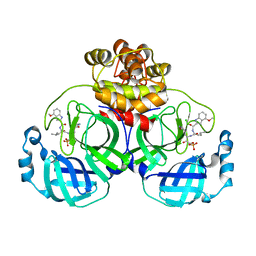 | | Crystal structure of SARS coronavirus main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of SARS coronavirus main protease in complex with PF07304814
To Be Published
|
|
5OYG
 
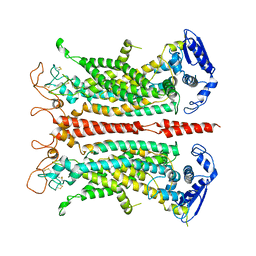 | | Structure of calcium-free mTMEM16A chloride channel at 4.06 A resolution | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
2MG8
 
 | |
4QR6
 
 | |
4RPQ
 
 | |
3KLQ
 
 | | Crystal Structure of the Minor Pilin FctB from Streptococcus pyogenes 90/306S | | Descriptor: | GLYCEROL, Putative pilus anchoring protein | | Authors: | Linke, C, Young, P.G, Bunker, R.D, Caradoc-Davies, T.T, Baker, E.N. | | Deposit date: | 2009-11-08 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring
J.Biol.Chem., 285, 2010
|
|
8TR9
 
 | | Cryo-EM structure of transglutaminase 2 bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase 2 | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational activation and inhibition of transglutaminase 2 determined by static and time resolved small-angle X-ray scattering and cryoelectron microscopy
To Be Published
|
|
4YS1
 
 | | Human Aldose Reductase complexed with a ligand with an IDD structure (2) at 1.07 A. | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Price for Opening the Transient Specificity Pocket in Human Aldose Reductase upon Ligand Binding: Structural, Thermodynamic, Kinetic, and Computational Analysis.
ACS Chem. Biol., 12, 2017
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
5OYB
 
 | | Structure of calcium-bound mTMEM16A chloride channel at 3.75 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Paulino, C, Kalienkova, V, Lam, K.M, Neldner, Y, Dutzler, R. | | Deposit date: | 2017-09-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM.
Nature, 552, 2017
|
|
4YU1
 
 | |
8ULG
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGB
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-05 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UFI
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
8UGS
 
 | | Cryo-EM structure of bovine phosphodiesterase 6 bound to cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Aplin, C, Cerione, R.A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6.
J.Biol.Chem., 300, 2023
|
|
4ES8
 
 | | Crystal Structure of the adhesin domain of Epf from Streptococcus pyogenes in P212121 | | Descriptor: | ACETATE ION, Epf, GLYCEROL, ... | | Authors: | Linke, C, Siemens, N, Kreikemeyer, B, Baker, E.N. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Extracellular Protein Factor Epf from Streptococcus pyogenes Is a Cell Surface Adhesin That Binds to Cells through an N-terminal Domain Containing a Carbohydrate-binding Module.
J.Biol.Chem., 287, 2012
|
|
