1U9Q
 
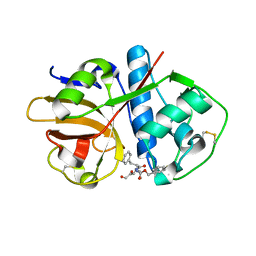 | | Crystal structure of cruzain bound to an alpha-ketoester | | Descriptor: | [1-(1-METHYL-4,5-DIOXO-PENT-2-ENYLCARBAMOYL)-2-PHENYL-ETHYL]-CARBAMIC ACID BENZYL ESTER, cruzipain | | Authors: | Lange, M, Weston, S.G, Cheng, H, Culliane, M, Fiorey, M.M, Grisostomi, C, Hardy, L.W, Hartstough, D.S, Pallai, P.V, Tilton, R.F, Baldino, C.M, Brinen, L.S, Engel, J.C, Choe, Y, Price, M.S, Craik, C.S. | | Deposit date: | 2004-08-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of alpha-keto-based inhibitors of cruzain, a cysteine protease implicated in Chagas disease
Bioorg.Med.Chem., 13, 2005
|
|
8BR6
 
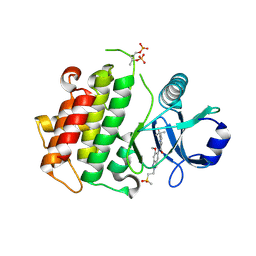 | | Discovery of IRAK4 Inhibitor 40 | | Descriptor: | ACETATE ION, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-methylsulfonylethyl)-1,3-dihydroindazol-5-yl]-6-(2-oxidanylpropan-2-yl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wengner, A.M, Guimond, N, Thaler, T, Platzek, J, Ewerspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.167 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8BR5
 
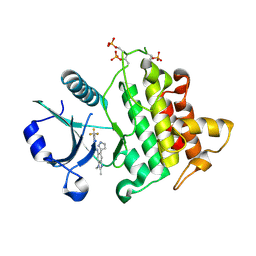 | | Discovery of IRAK4 Inhibitor 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[2,3-dimethyl-6-(1~{H}-pyrazol-5-yl)benzimidazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8BR7
 
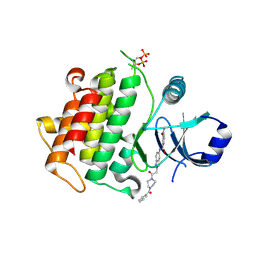 | | Discovery of IRAK4 Inhibitors BAY1834845 and BAY1830839 | | Descriptor: | 3-nitro-~{N}-[2-[2-oxidanylidene-2-[4-(phenylcarbonyl)piperazin-1-yl]ethyl]indazol-5-yl]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-11-22 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8ATL
 
 | | Discovery of IRAK4 Inhibitor 23 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[6-methoxy-2-(2-morpholin-4-yl-2-oxidanylidene-ethyl)indazol-5-yl]-6-[(1~{R})-2,2,2-tris(fluoranyl)-1-oxidanyl-ethyl]pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.464 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8ATN
 
 | | Discovery of IRAK4 Inhibitor 38 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-[3-methyl-2-(3-methyl-3-oxidanyl-butyl)-6-(2-oxidanylpropan-2-yl)benzimidazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
8ATB
 
 | | Discovery of IRAK4 Inhibitor 16 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 4, ~{N}-[6-ethoxy-2-[2-(4-methylpiperazin-1-yl)-2-oxidanylidene-ethyl]indazol-5-yl]-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Schafer, M, Bothe, U, Schmidt, N, Gunther, J, Nubbemeyer, R, Siebeneicher, H, Ring, S, Boemer, U, Peters, M, Denner, K, Himmel, H, Sutter, A, Terebesi, I, Lange, M, Wenger, A.M, Guimond, N, Thaler, T, Platzek, J, Eberspaecher, U, Steuber, H, Steinmeyer, A, Zollner, T.M. | | Deposit date: | 2022-08-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of IRAK4 Inhibitors BAY1834845 (Zabedosertib) and BAY1830839 .
J.Med.Chem., 67, 2024
|
|
6Y5B
 
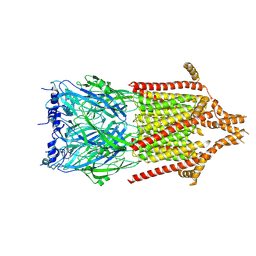 | | 5-HT3A receptor in Salipro (apo, asymmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6Y59
 
 | | 5-HT3A receptor in Salipro (apo, C5 symmetric) | | Descriptor: | 5-hydroxytryptamine receptor 3A | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
6Y5A
 
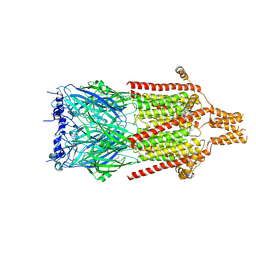 | | Serotonin-bound 5-HT3A receptor in Salipro | | Descriptor: | 5-hydroxytryptamine receptor 3A, SEROTONIN | | Authors: | Zhang, Y, Dijkman, P.M, Zou, R, Zandl-Lang, M, Sanchez, R.M, Eckhardt-Strelau, L, Koefeler, H, Vogel, H, Yuan, S, Kudryashev, M. | | Deposit date: | 2020-02-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Asymmetric opening of the homopentameric 5-HT 3A serotonin receptor in lipid bilayers.
Nat Commun, 12, 2021
|
|
8RDX
 
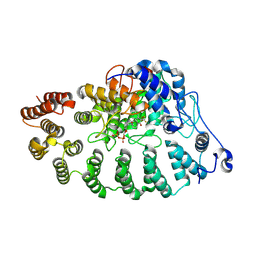 | | PGGtase I in complex with probe BAY-6092 | | Descriptor: | (5~{R})-5-(2-methoxyphenyl)-9-[(2~{R})-3,3,3-tris(fluoranyl)-2-methoxy-2-phenyl-propanoyl]-3,9-diazaspiro[5.5]undecan-2-one, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Steuber, H. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Discovery of YAP1/TAZ pathway inhibitors through phenotypic screening with potent anti-tumor activity via blockade of Rho-GTPase signaling.
Cell Chem Biol, 31, 2024
|
|
7LRD
 
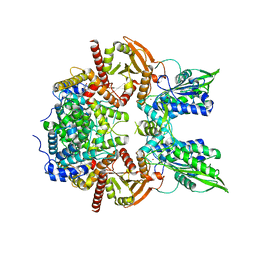 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
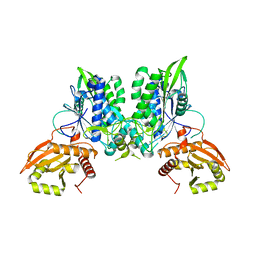
7LRE
 
 | |
7LRC
 
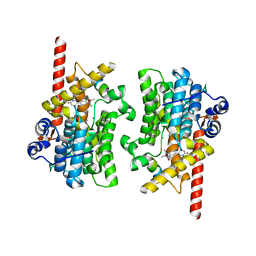 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
3O7A
 
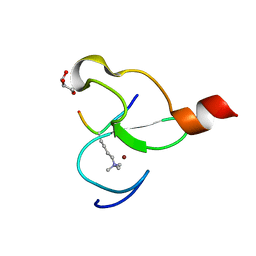 | | Crystal structure of PHF13 in complex with H3K4me3 | | Descriptor: | GLYCEROL, H3K4ME3 HISTONE 11MER-PEPTIDE, PHD finger protein 13 variant, ... | | Authors: | Bian, C.B, Lam, R, Xu, C, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-30 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.
Elife, 5, 2016
|
|
3O70
 
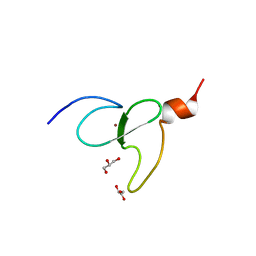 | | PHD-type zinc finger of human PHD finger protein 13 | | Descriptor: | GLYCEROL, PHD finger protein 13, ZINC ION | | Authors: | Lam, R, Bian, C.B, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.
Elife, 5, 2016
|
|
4BD3
 
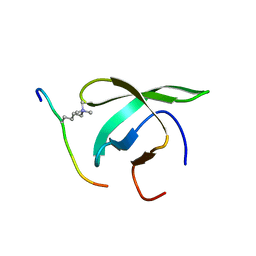 | |
7L28
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | Descriptor: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-12-16 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
 | |
7L27
 
 | |
7KWE
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to DNMDP | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
3KRR
 
 | | Crystal Structure of JAK2 complexed with a potent quinoxaline ATP site inhibitor | | Descriptor: | 8-[3,5-difluoro-4-(morpholin-4-ylmethyl)phenyl]-2-(1-piperidin-4-yl-1H-pyrazol-4-yl)quinoxaline, Tyrosine-protein kinase JAK2 | | Authors: | Tavares, G.A, Gerspacher, M, Kroemer, M, Scheufler, C. | | Deposit date: | 2009-11-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibition of Polycythemia by the Quinoxaline JAK2 Inhibitor NVP-BSK805
Mol.Cancer Ther., 9, 2010
|
|
1HII
 
 | |
1HIH
 
 | |
1EB4
 
 | |
