3KJX
 
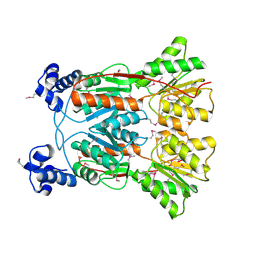 | |
4QTF
 
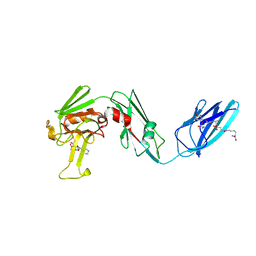 | |
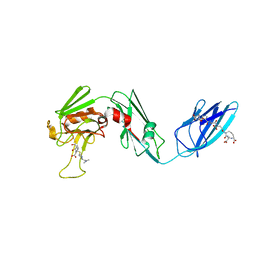
4QR7
 
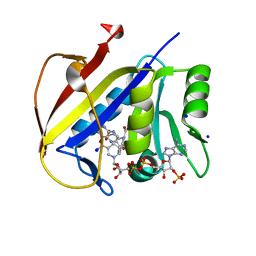
 | | Structure and specificity of L-D-Transpeptidase from Mycobacterium tuberculosis and antibiotic resistance: Calcium binding promotes dimer formation | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLCNAC(BETA1-4)-MURNAC(1,6-ANHYDRO)-L-ALA-GAMMA-D-GLU-MESO-A2PM-D-ALA, L,d-transpeptidase LdtB | | Authors: | Gokulan, K, Varughese, K.I. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-29 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure and specificity of L-D-Transpeptidase from Mycobacterium tuberculosis and antibiotic resistance: Calcium binding promotes dimer formation
To be Published
|
|
8E4F
 
 | | Crystal structure of dihydrofolate reductase (DHFR) from the filarial nematode W. bancrofti in complex with NADPH and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lange, K, Frey, K.M, Goodey, N.M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of dihydrofolate reductase from the filarial nematode W. bancrofti in complex with NADPH and folate.
Plos Negl Trop Dis, 17, 2023
|
|
1HKW
 
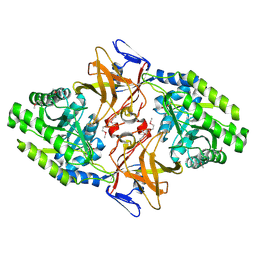 | | MYCOBACTERIUM DIAMINOPIMELATE DICARBOXYLASE (LysA) | | Descriptor: | DIAMINOPIMELATE DECARBOXYLASE, SULFATE ION | | Authors: | Gokulan, K, Rupp, B, Pavelka Jr, M.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-03-11 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Diaminopimelate Decarboxylase, an Essential Enzyme in Bacterial Lysine Biosynthesis
J.Biol.Chem., 278, 2003
|
|
1HKV
 
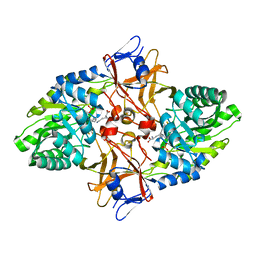 | | mycobacterium diaminopimelate dicarboxylase (lysa) | | Descriptor: | DIAMINOPIMELATE DECARBOXYLASE, LYSINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gokulan, K, Rupp, B, Pavelka Jr, M.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-03-11 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Diaminopimelate Decarboxylase, an Essential Enzyme in Bacterial Lysine Biosynthesis
J.Biol.Chem., 278, 2003
|
|
8STT
 
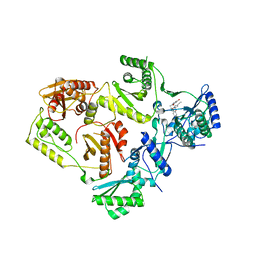 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STR
 
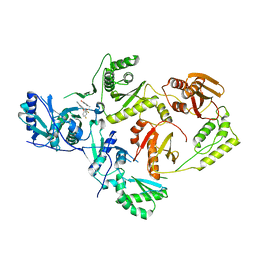 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Chan, A.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STP
 
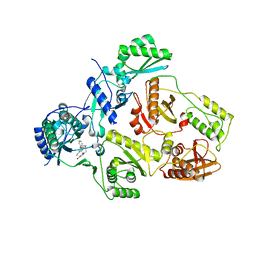 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Frey, K.M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STQ
 
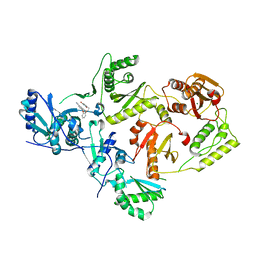 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Frey, K.M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STS
 
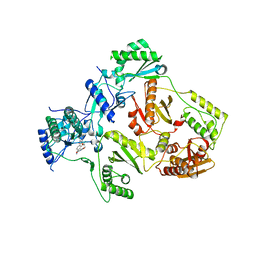 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STU
 
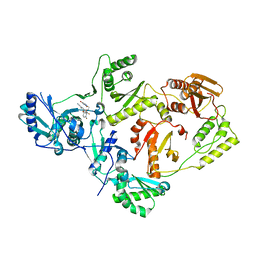 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-6-fluoroindolizine-2-carbonitrile (JLJ578), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
8STV
 
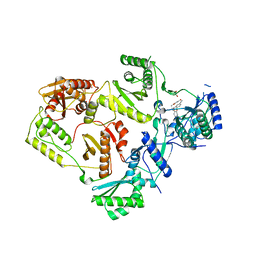 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) variant in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-2-naphthonitrile (JLJ600), a non-nucleoside inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}naphthalene-2-carbonitrile, MAGNESIUM ION, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
1YNK
 
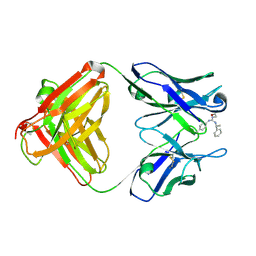 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeteners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-[((R)-{[4-(AMINOMETHYL)PHENYL]AMINO}{[(1R)-1-PHENYLETHYL]AMINO}METHYL)AMINO]ETHANE-1,1-DIOL, Ig gamma heavy chain, immunoglobulin kappa light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
1YNL
 
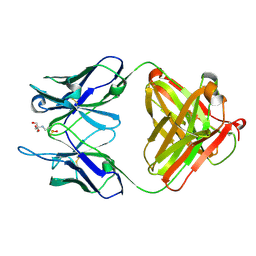 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeterners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Ig gamma heavy chain, Ig gamma light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
3H5T
 
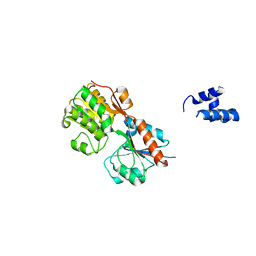 | |
3GPV
 
 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis | | Descriptor: | Transcriptional regulator, MerR family | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a transcriptional regulator, MerR family from Bacillus thuringiensis
To be Published
|
|
3KZG
 
 | |
3GYB
 
 | |
3GRC
 
 | |
3HH0
 
 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus cereus | | Descriptor: | Transcriptional regulator, MerR family | | Authors: | Palani, K, Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-14 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of a transcriptional regulator, MerR family from Bacillus cereus
To be Published
|
|
3HD5
 
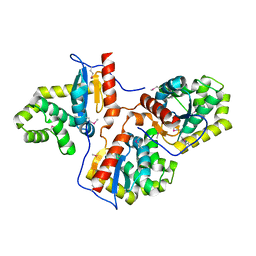 | |
3LHL
 
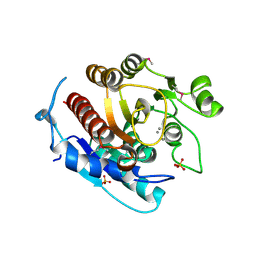 | | Crystal structure of a putative agmatinase from Clostridium difficile | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative agmatinase from Clostridium difficile
To be Published
|
|
3LOP
 
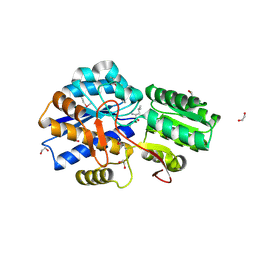 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
3LTO
 
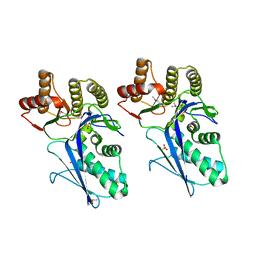 | |
