4LLS
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP, and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with IPP, GSPP and calcium bound in active site
To be Published
|
|
4LLT
 
 | | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, Geranyltranstransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-09 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a farnesyl diphosphate synthase from Roseobacter denitrificans OCh 114, target EFI-509393, with two IPP and calcium bound in active site
To be Published
|
|
3SGL
 
 | |
7EQ5
 
 | | Plant growth-promoting factor YxaL mutant from Bacillus velezensis - T175W/W215G | | Descriptor: | Membrane associated protein kinase with beta-propeller domain, pyrrolo-quinoline quinone beta-propeller repeat | | Authors: | Kim, J, Ha, N.-C. | | Deposit date: | 2021-04-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the plant growth-promoting factor YxaL from the rhizobacterium Bacillus velezensis and its application to protein engineering.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7EVF
 
 | |
8GUY
 
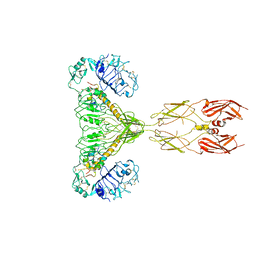 | | human insulin receptor bound with two insulin molecules | | Descriptor: | Insulin A chain, Insulin, isoform 2, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ3
 
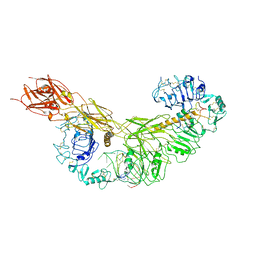 | | human insulin receptor bound with A43 DNA aptamer and insulin | | Descriptor: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ4
 
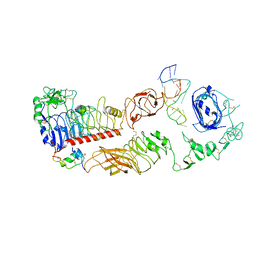 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ6
 
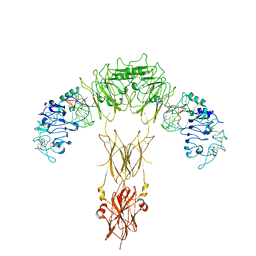 | | human insulin receptor bound with A62 DNA aptamer | | Descriptor: | IR-A62 aptamer, Isoform Short of Insulin receptor | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
9JG0
 
 | | Cryo-EM structure of neuropeptide FF receptor 2 in the ligand-free state with BRIL fusion, anti-BRIL Fab, and nanobody | | Descriptor: | Anti-BRIL fab heavy chain, Anti-BRIL fab light chain, Anti-fab nanobody, ... | | Authors: | Kim, J, Choi, H.-J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural insights into the selective recognition of RF-amide peptides by neuropeptide FF receptor 2.
Embo Rep., 26, 2025
|
|
9JFY
 
 | | Cryo-EM structure of Neuropeptide FF receptor 2 in complex with hNPSF and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kim, J, Choi, H.-J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural insights into the selective recognition of RF-amide peptides by neuropeptide FF receptor 2.
Embo Rep., 26, 2025
|
|
8K6X
 
 | | Crystal structure of E.coli Cyanase complex with cyanate and bicarbonate | | Descriptor: | CARBONATE ION, Cyanate hydratase, SULFATE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of Escherichia coli cyanase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8K6U
 
 | |
8K6H
 
 | |
8K6S
 
 | |
8K6G
 
 | | Crystal structure of E.coli Cyanase | | Descriptor: | Cyanate hydratase, SULFATE ION | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural mechanism of Escherichia coli cyanase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5HZ2
 
 | | Crystal structure of PhaC1 from Ralstonia eutropha | | Descriptor: | GLYCEROL, Poly-beta-hydroxybutyrate polymerase, SULFATE ION | | Authors: | Kim, J, Kim, K.-J. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.
Biotechnol J, 12, 2017
|
|
9IPY
 
 | | Structure of JR14a-bound human C3aR | | Descriptor: | (2~{S})-5-[bis(azanyl)methylideneamino]-2-[[5-[bis(4-chlorophenyl)methyl]-3-methyl-thiophen-2-yl]carbonylamino]pentanoic acid, C3a anaphylatoxin chemotactic receptor,Soluble cytochrome b562 | | Authors: | Kim, J, Ko, S, Choi, H.-J. | | Deposit date: | 2024-07-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into small-molecule agonist recognition and activation of complement receptor C3aR.
Embo J., 44, 2025
|
|
9IPV
 
 | | Structure of JR14a-C3aR-Gi-scFv16 complex | | Descriptor: | (2~{S})-5-[bis(azanyl)methylideneamino]-2-[[5-[bis(4-chlorophenyl)methyl]-3-methyl-thiophen-2-yl]carbonylamino]pentanoic acid, C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kim, J, Ko, S, Choi, H.-J. | | Deposit date: | 2024-07-11 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into small-molecule agonist recognition and activation of complement receptor C3aR.
Embo J., 44, 2025
|
|
9ISI
 
 | | Structure of human C3aR in apo state | | Descriptor: | C3a anaphylatoxin chemotactic receptor,Soluble cytochrome b562 | | Authors: | Kim, J, Ko, S, Choi, H.-J. | | Deposit date: | 2024-07-17 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into small-molecule agonist recognition and activation of complement receptor C3aR.
Embo J., 44, 2025
|
|
9JT7
 
 | | SFX reaction state structure (0-60min) of alanine racemase | | Descriptor: | ALANINE, Alanine racemase 2, CHLORIDE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2024-10-02 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the reaction dynamics of alanine racemase using serial femtosecond crystallography.
Sci Rep, 14, 2024
|
|
8H26
 
 | |
1MIO
 
 | | X-RAY CRYSTAL STRUCTURE OF THE NITROGENASE MOLYBDENUM-IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Kim, J, Woo, D, Rees, D.C. | | Deposit date: | 1993-03-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0-A resolution.
Biochemistry, 32, 1993
|
|
8JQV
 
 | |
