7MWR
 
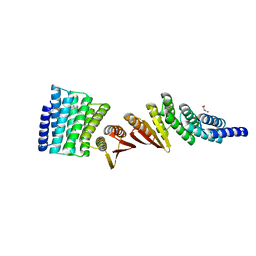 | | Structure of De Novo designed beta sheet heterodimer LHD101A53/B4 | | Descriptor: | LHD101A54, LHD101B4, MALONATE ION | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
7MWQ
 
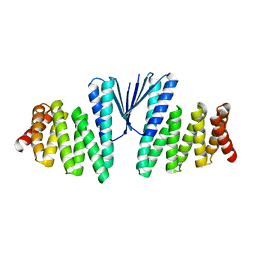 | | Structure of De Novo designed beta sheet heterodimer LHD29A53/B53 | | Descriptor: | LHD29A53, LHD29B53 | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Praetorius, F, Baker, D. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
8T5E
 
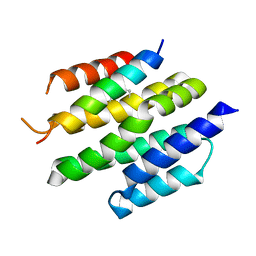 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Bcl-2-like protein 11, Bim_fulldiff | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8T5F
 
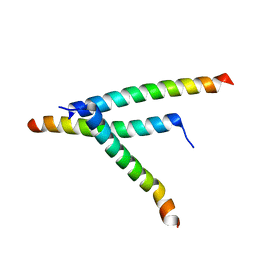 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Parathyroid hormone | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8T6E
 
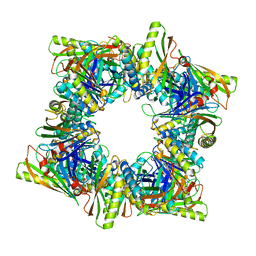 | | Crystal structure of T33-28.3: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-28.3: A, T33-28.3: B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T6C
 
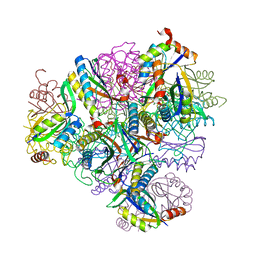 | | Crystal structure of T33-18.2: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-18.2 : A, T33-18.2 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-15 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7RMY
 
 | | De Novo designed tunable protein pockets, D_3-337 | | Descriptor: | De Novo designed tunable homodimer, D_3-337 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RMX
 
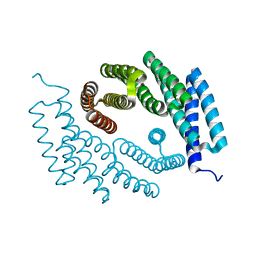 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8T6N
 
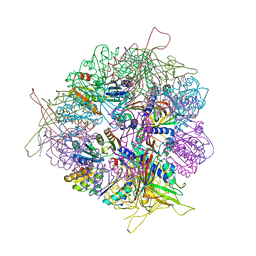 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-27.1 : A, T33-27.1 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7KXS
 
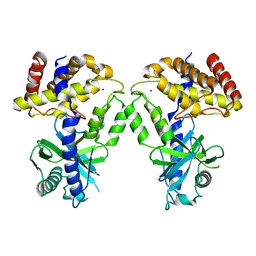 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7LZ3
 
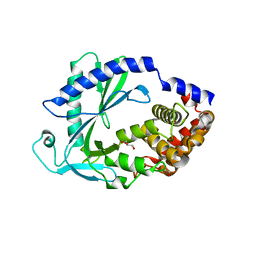 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6WMK
 
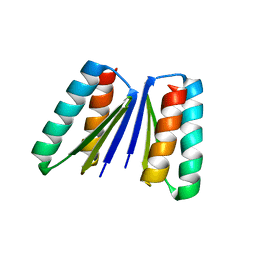 | | Crystal structure of beta sheet heterodimer LHD29 | | Descriptor: | Beta sheet heterodimer LHD29 - Chain A, Beta sheet heterodimer LHD29 - Chain B | | Authors: | Bera, A.K, Sahtoe, D.D, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2020-04-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reconfigurable asymmetric protein assemblies through implicit negative design.
Science, 375, 2022
|
|
8GJI
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | GCG binder, Glucagon | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GJG
 
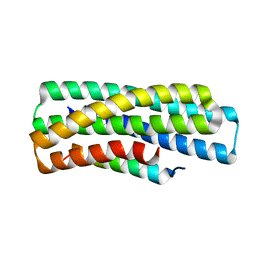 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8G9K
 
 | |
8GA6
 
 | |
8GA7
 
 | |
8UTK
 
 | | IL-23R minibinder - 23R-B04dslf02IB | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 23R-B04dslf02IB, ... | | Authors: | Bera, A.K, Berger, S.A, Kang, A, Baker, D. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Preclinical proof of principle for orally delivered Th17 antagonist miniproteins.
Cell, 187, 2024
|
|
8VEJ
 
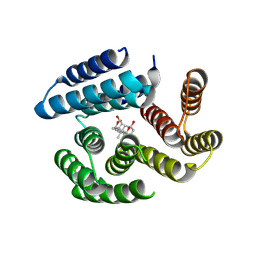 | | De novo designed cholic acid binder: CHD_buttress | | Descriptor: | CHD_buttress, CHOLIC ACID | | Authors: | Bera, A.K, Tran, L, Kang, A, Baker, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Binding and sensing diverse small molecules using shape-complementary pseudocycles.
Science, 385, 2024
|
|
6V67
 
 | | Apo Structure of the De Novo PD-1 Binding Miniprotein GR918.2 | | Descriptor: | PD-1 Binding Miniprotein GR918.2 | | Authors: | Bick, M.J, Bryan, C.M, Baker, D, Dimaio, F, Kang, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Computational design of a synthetic PD-1 agonist.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8UGC
 
 | |
8UZL
 
 | | Designed Transmembrane beta-barrel- TMB10_163 | | Descriptor: | Designed Transmembrane beta-barrel TMB10_163, HEXANE-1,6-DIOL | | Authors: | Bera, A.K, Lemma, S.B, Kang, A, Baker, D. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sculpting conducting nanopore size and shape through de novo protein design.
Science, 385, 2024
|
|
8VD6
 
 | |
8EOZ
 
 | |
8GL3
 
 | |
