2JX8
 
 | | Solution structure of hPCIF1 WW domain | | Descriptor: | Phosphorylated CTD-interacting factor 1 | | Authors: | Kouno, T, Iwamoto, Y, Hirose, Y, Aizawa, T, Demura, M, Kawano, K, Ohkuma, Y, Mizuguchi, M. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, and 15N resonance assignments of hPCIF1 WW domain
To be Published
|
|
8K9O
 
 | |
6K55
 
 | | Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide | | Descriptor: | Glucanase, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6K54
 
 | | Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with trisaccharide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Novel hyperthermophilic cellobiohydrolase II isolated from hot spring microbial community
To Be Published
|
|
6K53
 
 | | A variant of metagenome-derived GH6 cellobiohydrolase, HmCel6A (P88S/L230F/F414S) | | Descriptor: | CITRATE ANION, GH6 cellobiohydrolase, HMCEL6A, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from a hot spring metagenomic data
To Be Published
|
|
6K52
 
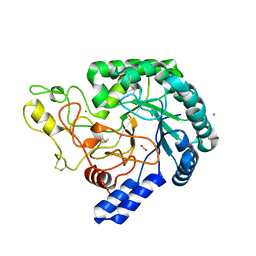 | | Hyperthermophilic GH6 cellobiohydrolase (HmCel6A) from the microbial flora of a Japanese hot spring | | Descriptor: | ACETATE ION, CALCIUM ION, GH6 cellobiohydrolase, ... | | Authors: | Baba, S, Takeda, M, Okuma, J, Hirose, Y, Nishimura, A, Takata, M, Oda, K, Shibata, D, Kondo, Y, Kumasaka, T. | | Deposit date: | 2019-05-28 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68000138 Å) | | Cite: | A hyperthermophilic cellobiohydrolase mined from a hot spring metagenomic data
To Be Published
|
|
6M5B
 
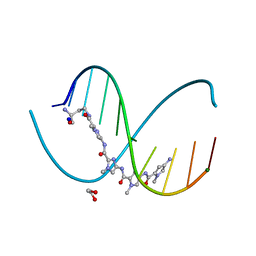 | | X-ray crystal structure of cyclic-PIP and DNA complex in a reverse binding orientation | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[(4-azanyl-1-methyl-pyrrol-2-yl)carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-~{N}-[2-[[(3~{S})-3-azanyl-4-oxidanylidene-butyl]carbamoyl]-1-methyl-imidazol-4-yl]-1-methyl-imidazole-2-carboxamide, DNA (5'-D(*CP*(CBR)P*AP*GP*GP*CP*CP*TP*GP*G)-3'), ... | | Authors: | Abe, K, Hirose, Y, Eki, H, Takeda, K, Bando, T, Endo, M, Sugiyama, H. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | X-ray Crystal Structure of a Cyclic-PIP-DNA Complex in the Reverse-Binding Orientation.
J.Am.Chem.Soc., 142, 2020
|
|
7VEB
 
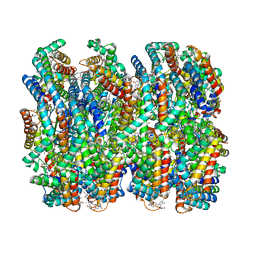 | | Phycocyanin rod structure of cyanobacterial phycobilisome | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Kawakami, K, Hamaguchi, T, Hirose, Y, Kosumi, D, Miyata, M, Kamiya, N, Yonekura, K. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Nat Commun, 13, 2022
|
|
7VEA
 
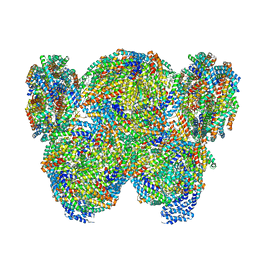 | | Pentacylindrical allophycocyanin core from Thermosynechococcus vulcanus | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Kawakami, K, Hamaguchi, T, Hirose, Y, Kosumi, D, Miyata, M, Kamiya, N, Yonekura, K. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Nat Commun, 13, 2022
|
|
7CKV
 
 | | Crystal structure of Cyanobacteriochrome GAF domain in Pr state | | Descriptor: | MAGNESIUM ION, PHYCOCYANOBILIN, RcaE | | Authors: | Nagae, T, Koizumi, T, Hirose, Y, Mishima, M. | | Deposit date: | 2020-07-19 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XAR
 
 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | Deposit date: | 2022-03-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
7SAF
 
 | |
7SAY
 
 | |
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
8WEY
 
 | | PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137) | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{R} )-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nakajima, Y, Kawakami, K, Yonekura, K, Shen, J.R, Nagao, R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | The structure of PSI-LHCI from Cyanidium caldarium provides evolutionary insights into conservation and diversity of red-lineage LHCs.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6IRZ
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, PDX1 C-terminal-inhibiting factor 1, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRV
 
 | |
6IS0
 
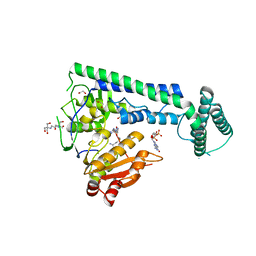 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRY
 
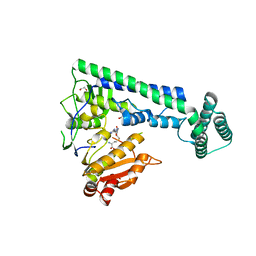 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IRX
 
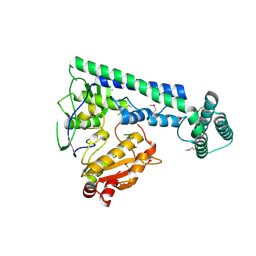 | |
6IRW
 
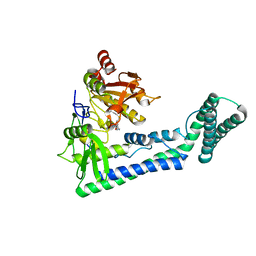 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
8WU5
 
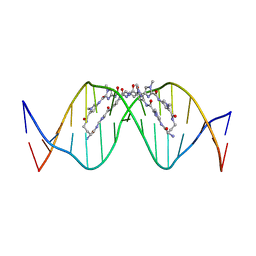 | | The complex of CAG repeat sequence-specific binding cPIP and dsDNA with A-A mismatch | | Descriptor: | (1^2Z,4^2Z,11^2Z,14^2Z,22^2Z,25^2Z,32^2Z,35^2Z,19R,40R)-1^1,4^1,11^1,14^1,22^1,25^1,32^1,35^1-octamethyl-2,5,9,12,15,20,23,26,30,33,36,41-dodecaoxo-1^1H,4^1H,11^1H,14^1H,22^1H,25^1H,32^1H,35^1H-3,6,10,13,16,21,24,27,31,34,37,42-dodecaaza-1(2,4),11,22,32(4,2)-tetraimidazola-4,14,25,35(4,2)-tetrapyrrolacyclodotetracontaphane-19,40-diaminium, DNA (5'-D(*GP*CP*(CBR)P*GP*AP*GP*CP*AP*GP*CP*AP*CP*GP*GP*C)-3') | | Authors: | Abe, K, Takeda, K, Sugiyama, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of a Complex of a CAG/CTG Repeat Sequence-Specific Binding Molecule and A-A-Mismatch-Containing DNA.
Jacs Au, 4, 2024
|
|
5GGG
 
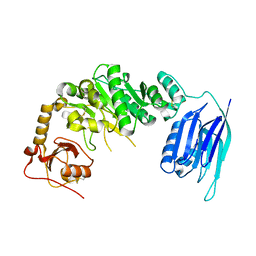 | |
5GGN
 
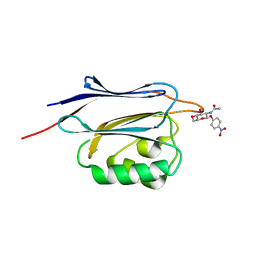 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGJ
 
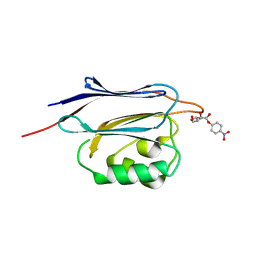 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-alpha-pNP | | Descriptor: | 4-nitrophenyl alpha-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
