6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EZ8
 
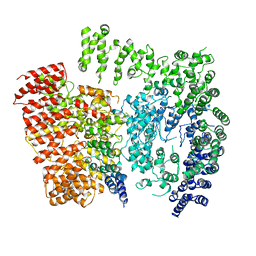 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
3J2H
 
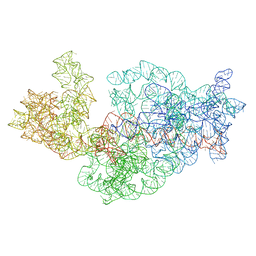 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2B
 
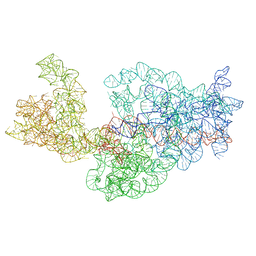 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2A
 
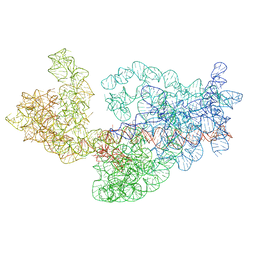 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
2WK3
 
 | |
2YKR
 
 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1GIU
 
 | | A TRICHOSANTHIN(TCS) MUTANT(E85R) COMPLEX STRUCTURE WITH ADENINE | | Descriptor: | ADENINE, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Guo, Q, Liu, Y, Dong, Y, Rao, Z. | | Deposit date: | 2001-03-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate binding and catalysis in trichosanthin occur in different sites as revealed by the complex structures of several E85 mutants.
Protein Eng., 16, 2003
|
|
1SK6
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin, 3',5' cyclic AMP (cAMP), and pyrophosphate | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Shen, Y, Zhukovskaya, N.L, Tang, W.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate.
J.Biol.Chem., 279, 2004
|
|
1YRU
 
 | |
1YRT
 
 | | Crystal Structure analysis of the adenylyl cyclaes catalytic domain of adenylyl cyclase toxin of Bordetella pertussis in presence of c-terminal calmodulin | | Descriptor: | Bifunctional hemolysin-adenylate cyclase, CALCIUM ION, Calmodulin | | Authors: | Guo, Q, Shen, Y, Lee, Y.S, Gibbs, C.S, Mrksich, M, Tang, W.J. | | Deposit date: | 2005-02-04 | | Release date: | 2006-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin.
Embo J., 24, 2005
|
|
1ZOT
 
 | | crystal structure analysis of the CyaA/C-Cam with PMEAPP | | Descriptor: | (ADENIN-9-YL-ETHOXYMETHYL)-HYDROXYPHOSPHINYL-DIPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2005-05-13 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin.
Embo J., 24, 2005
|
|
2COL
 
 | | Crystal structure analysis of CyaA/C-Cam with Pyrophosphate | | Descriptor: | Bifunctional hemolysin-adenylate cyclase, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Tang, W.J, Shen, Y. | | Deposit date: | 2005-05-18 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin
Embo J., 24, 2005
|
|
3E4Z
 
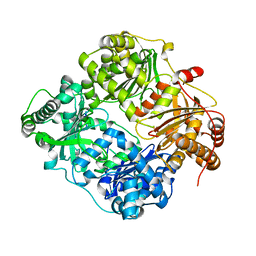 | | Crystal structure of human insulin degrading enzyme in complex with insulin-like growth factor II | | Descriptor: | Insulin-degrading enzyme, Insulin-like growth factor II, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
3E50
 
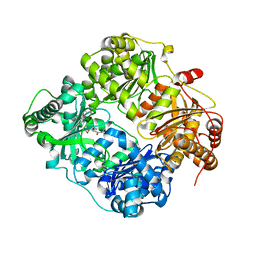 | | Crystal structure of human insulin degrading enzyme in complex with transforming growth factor-alpha | | Descriptor: | Insulin-degrading enzyme, Protransforming growth factor alpha, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
6EPC
 
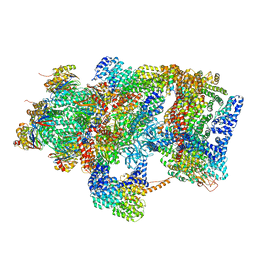 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
8TTU
 
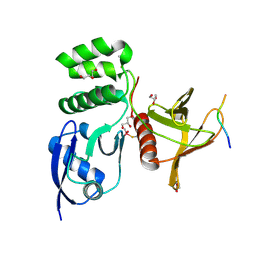 | | Structure of SNX27 FERM complexed with Fam21A repeat 19 (1261 - 1274) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fam21A repeat 19 peptide, GLYCEROL, ... | | Authors: | Guo, Q, Chen, K.-E, Collins, B.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for coupling of the WASH subunit FAM21 with the endosomal SNX27-Retromer complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TTV
 
 | |
8TTT
 
 | | Structure of SNX27 FERM complexed with Fam21A repeat 15 (1124-1140) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fam21A repeat 15 peptide, GLYCEROL, ... | | Authors: | Guo, Q, Chen, K.-E, Collins, B.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for coupling of the WASH subunit FAM21 with the endosomal SNX27-Retromer complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5DNA
 
 | | Crystal structure of Candida boidinii formate dehydrogenase | | Descriptor: | FORMATE DEHYDROGENASE, SULFATE ION | | Authors: | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
3HGZ
 
 | | Crystal structure of human insulin-degrading enzyme in complex with amylin | | Descriptor: | Insulin-degrading enzyme, Islet amyloid polypeptide, ZINC ION | | Authors: | Guo, Q, Bian, Y, Tang, W.J. | | Deposit date: | 2009-05-14 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
5DN9
 
 | | Crystal structure of Candida boidinii formate dehydrogenase complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, FDH, ... | | Authors: | Guo, Q, Gakhar, L, Wichersham, K, Francis, K, Vardi-Kilshtain, A, Major, D.T, Cheatum, C.M, Kohen, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Kinetic Studies of Formate Dehydrogenase from Candida boidinii.
Biochemistry, 55, 2016
|
|
3J2G
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
