7QEL
 
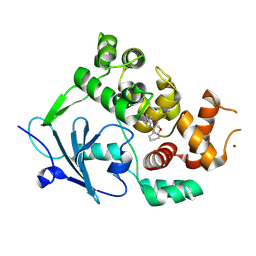 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247 | | Descriptor: | (2~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)spiro[2.3]hexane-2-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247
To Be Published
|
|
6TWO
 
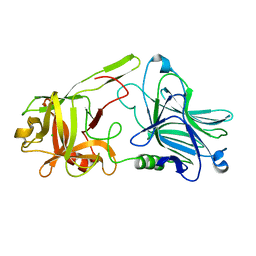 | | Binding domain of BoNT/A6 | | Descriptor: | Bont/A1, CHLORIDE ION | | Authors: | Davies, J.R, Britton, A, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
6TWP
 
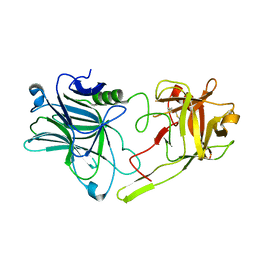 | | Binding domain of BoNT/A5 | | Descriptor: | Botulinum neurotoxin A5, CHLORIDE ION | | Authors: | Davies, J.R, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
7Z5R
 
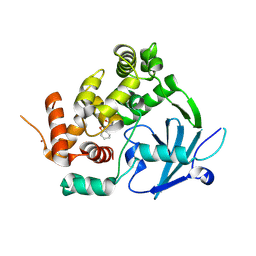 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035 | | Descriptor: | (7~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)bicyclo[4.2.0]octa-1,3,5-triene-7-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035
To Be Published
|
|
7ZC7
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941
To Be Published
|
|
7ZG3
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-[(1~{S})-1,2,2-trimethylcyclopropyl]pyrrolo[1,2-c]pyrimidine-3-carboxamide | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-04-01 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228
To Be Published
|
|
6ZVN
 
 | |
6ZVM
 
 | | Botulinum neurotoxin B2 binding domain in complex with GD1a | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin | | Authors: | Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of Botulinum Neurotoxin Subtype B2 Binding to Its Receptors.
Toxins, 12, 2020
|
|
6F0P
 
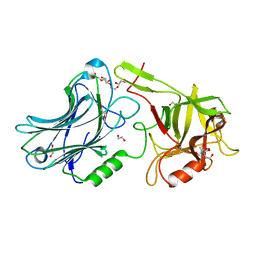 | | Botulinum neurotoxin A4 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, J.R, Rees, J, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
6F0O
 
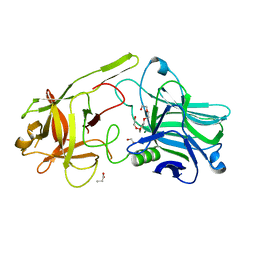 | | Botulinum neurotoxin A3 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, Bontoxilysin A, PENTAETHYLENE GLYCOL, ... | | Authors: | Davies, J.R, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
5MK6
 
 | |
5MK8
 
 | |
5MK7
 
 | |
4G9Y
 
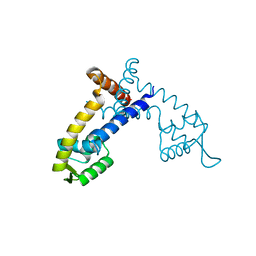 | | Crystal Structure of the PcaV transcriptional regulator from Streptomyces coelicolor | | Descriptor: | GLYCEROL, PcaV transcriptional regulator | | Authors: | Brown, B.L, Davis, J.R, Sello, J.K, Page, R. | | Deposit date: | 2012-07-24 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors.
Nucleic Acids Res., 41, 2013
|
|
4FHT
 
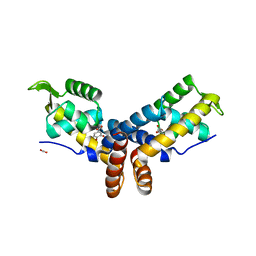 | | Crystal Structure of the PcaV transcriptional regulator from Streptomyces coelicolor in complex with its natural ligand | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, PcaV transcriptional regulator | | Authors: | Brown, B.L, Davis, J.R, Sello, J.K, Page, R. | | Deposit date: | 2012-06-06 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors.
Nucleic Acids Res., 41, 2013
|
|
4IGB
 
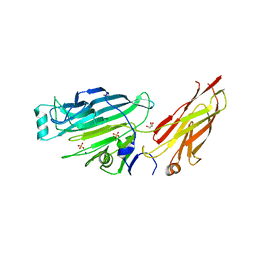 | | Crystal structure of the N-terminal domain of the Streptococcus gordonii adhesin Sgo0707 | | Descriptor: | ACETATE ION, GLYCEROL, LPXTG cell wall surface protein, ... | | Authors: | Nylander, A, Svensater, G, Senadheera, D.B, Cvitkovitch, D.G, Davies, J.R, Persson, K. | | Deposit date: | 2012-12-17 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Functional Analysis of the N-terminal Domain of the Streptococcus gordonii Adhesin Sgo0707
Plos One, 8, 2013
|
|
5BQN
 
 | | Crystal structure of the LHn fragment of botulinum neurotoxin type D, mutant H233Y E230Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Botulinum neurotoxin type D,Botulinum neurotoxin type D | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
5BQM
 
 | | Crystal structure of SXN101959, a Clostridium botulinum neurotoxin type D derivative and targeted secretion inhibitor | | Descriptor: | Botulinum neurotoxin type D, Somatoliberin,Botulinum neurotoxin type D, ZINC ION | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
7QFQ
 
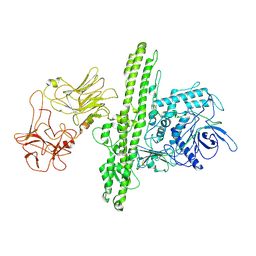 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7QFP
 
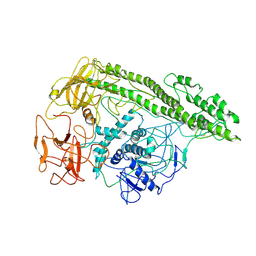 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7AZ0
 
 | |
7AYY
 
 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
 | |
7OVW
 
 | | Binding domain of botulinum neurotoxin E in complex with GD1a | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type E | | Authors: | Masuyer, G, Stenmark, P. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Ganglioside Receptor Recognition by Botulinum Neurotoxin Serotype E.
Int J Mol Sci, 22, 2021
|
|
