1BVE
 
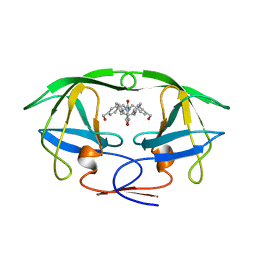 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR, 28 STRUCTURES | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1BVG
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
1RCH
 
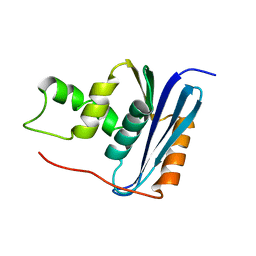 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
1C9F
 
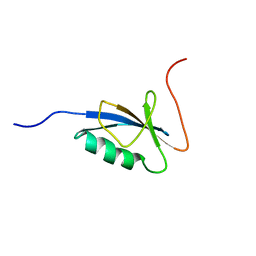 | |
1IRZ
 
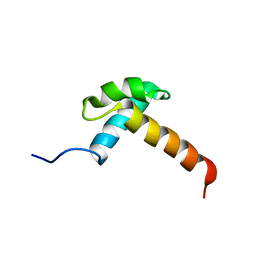 | |
6LK7
 
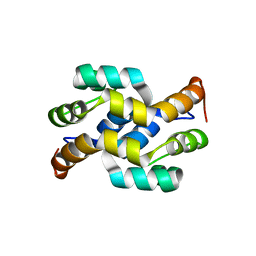 | | crystal structure of Os1348 from Pseudomonas sp. Os17 | | Descriptor: | Nitrile hydratase, alpha chain | | Authors: | Takeuchi, K, Tsuchiya, W, Fujimoto, Z, Yamada, K, Someya, N, Yamazaki, T. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Discovery of an Antibiotic-Related Small Protein of Biocontrol Strain Pseudomonas sp. Os17 by a Genome-Mining Strategy.
Front Microbiol, 11, 2020
|
|
2RS2
 
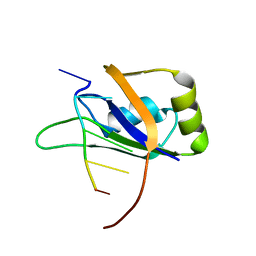 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | Deposit date: | 2011-06-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
7D2O
 
 | |
6M0Q
 
 | | Hydroxylamine oxidoreductase from Nitrosomonas europaea | | Descriptor: | Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
6M0P
 
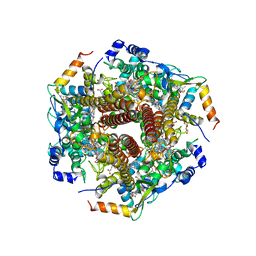 | | Hydroxylamine oxidoreductase in complex with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
8XZ2
 
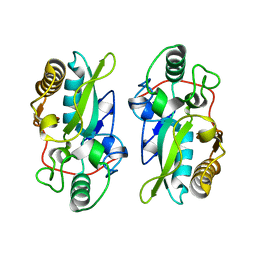 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
1AYG
 
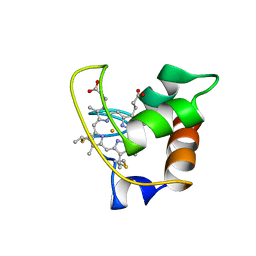 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
2RNN
 
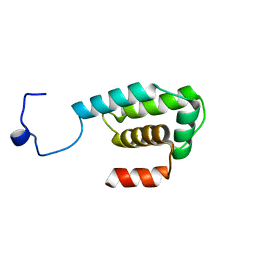 | |
2RNO
 
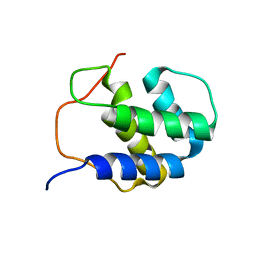 | |
2RQF
 
 | | Solution structure of juvenile hormone binding protein from silkworm in complex with JH III | | Descriptor: | Hemolymph juvenile hormone binding protein, methyl (2E,6E)-9-[(2R)-3,3-dimethyloxiran-2-yl]-3,7-dimethylnona-2,6-dienoate | | Authors: | Suzuki, R, Fujimoto, Z, Shiotsuki, T, Momma, M, Tase, A, Yamazaki, T. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-05 | | Last modified: | 2013-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of JH delivery in hemolymph by JHBP of silkworm, Bombyx mori
Sci Rep, 1, 2011
|
|
2PLE
 
 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
2PLD
 
 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
1F2R
 
 | | NMR STRUCTURE OF THE HETERODIMERIC COMPLEX BETWEEN CAD DOMAINS OF CAD AND ICAD | | Descriptor: | CASPASE-ACTIVATED DNASE, INHIBITOR OF CASPASE-ACTIVATED DNASE | | Authors: | Otomo, T, Sakahira, H, Uegaki, K, Nagata, S, Yamazaki, T. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the heterodimeric complex between CAD domains of CAD and ICAD.
Nat.Struct.Biol., 7, 2000
|
|
1DCJ
 
 | | SOLUTION STRUCTURE OF YHHP, A NOVEL ESCHERICHIA COLI PROTEIN IMPLICATED IN THE CELL DIVISION | | Descriptor: | YHHP PROTEIN | | Authors: | Katoh, E, Hatta, T, Shindo, H, Mizuno, T, Yamazaki, T. | | Deposit date: | 1999-11-05 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division.
J.Mol.Biol., 304, 2000
|
|
3AOS
 
 | | Crystal structure of juvenile hormone binding protein from silkworm in complex with JH II | | Descriptor: | Hemolymph juvenile hormone binding protein, methyl (2E,6E)-9-[(2R,3S)-3-ethyl-3-methyloxiran-2-yl]-3,7-dimethylnona-2,6-dienoate | | Authors: | Fujimoto, Z, Suzuki, R, Shiotsuki, T, Momma, M, Yamazaki, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanism of JH delivery in hemolymph by JHBP of silkworm, Bombyx mori
Sci Rep, 1, 2011
|
|
3AOT
 
 | | Crystal structure of juvenile hormone binding protein from silkworm in its apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hemolymph juvenile hormone binding protein, TETRAETHYLENE GLYCOL, ... | | Authors: | Fujimoto, Z, Suzuki, R, Shiotsuki, T, Momma, M, Yamazaki, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanism of JH delivery in hemolymph by JHBP of silkworm, Bombyx mori.
Sci Rep, 1, 2011
|
|
3A1Z
 
 | | Crystal structure of juvenile hormone binding protein from silkworm | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hemolymph juvenile hormone binding protein, ZINC ION | | Authors: | Suzuki, R, Fujimoto, Z, Shiotsuki, T, Momma, M, Tase, A, Yamazaki, T. | | Deposit date: | 2009-04-27 | | Release date: | 2010-04-28 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of silkworm Bombyx mori JHBP in complex with 2-methyl-2,4-pentanediol: plasticity of JH-binding pocket and ligand-induced conformational change of the second cavity in JHBP
Plos One, 8, 2013
|
|
1C4L
 
 | | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | | Descriptor: | RNA (5'-R(*CP*CP*UP*GP*CP*GP*UP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*UP*CP*AP*GP*G)-3') | | Authors: | Tanaka, Y, Kojima, C, Yamazaki, T, Kodama, T.S, Yasuno, K, Miyashita, S, Ono, A.M, Ono, A.S, Kainosho, M, Kyogoku, Y. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA duplex including a C-U base pair.
Biochemistry, 39, 2000
|
|
1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
