3SLU
 
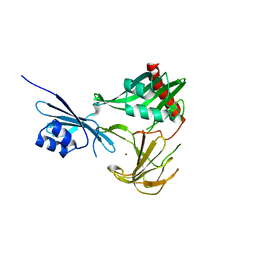 | | Crystal structure of NMB0315 | | Descriptor: | M23 peptidase domain protein, NICKEL (II) ION | | Authors: | Shen, Y, Wang, X, Yang, X, Xu, H. | | Deposit date: | 2011-06-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of outer membrane protein NMB0315 from Neisseria meningitidis.
Plos One, 6, 2011
|
|
5T4D
 
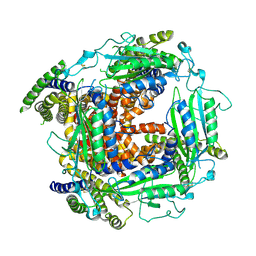 | | Cryo-EM structure of Polycystic Kidney Disease protein 2 (PKD2), residues 198-703 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hPKD:198-703, Polycystin-2 | | Authors: | Shen, P.S, Yang, X, DeCaen, P.G, Liu, X, Bulkley, D, Clapham, D.E, Cao, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs.
Cell, 167, 2016
|
|
4ZM6
 
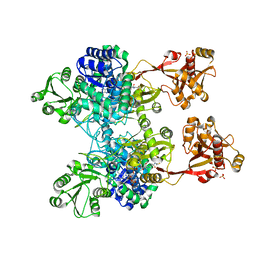 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ROU
 
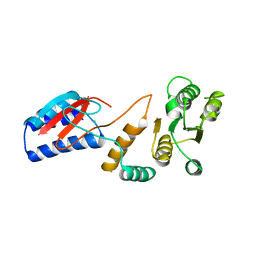 | |
8DQL
 
 | | CryoEM structure of IglD | | Descriptor: | Secretion system protein | | Authors: | Liu, X, Clemens, D, Lee, B, Yang, X, Zhou, H, Horwitz, M. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic Structure of IglD Demonstrates Its Role as a Component of the Baseplate Complex of the Francisella Type VI Secretion System.
Mbio, 13, 2022
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | Descriptor: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | Authors: | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | Deposit date: | 2001-10-24 | | Release date: | 2003-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
3TUO
 
 | |
1NBS
 
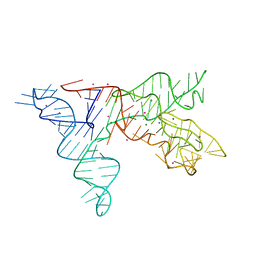 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | Descriptor: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | Authors: | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | Deposit date: | 2002-12-03 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
3UIT
 
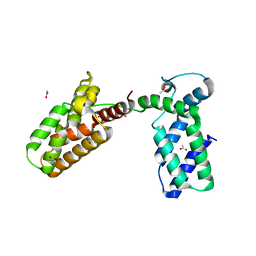 | | Overall structure of Patj/Pals1/Mals complex | | Descriptor: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | Authors: | Zhang, J, Yang, X, Long, J, Shen, Y. | | Deposit date: | 2011-11-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
4Q2J
 
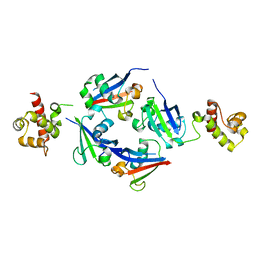 | | A novel structure-based mechanism for DNA-binding of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Wang, Z, Long, J, Yang, X, Shen, Y. | | Deposit date: | 2014-04-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Crystal structure of the ubiquitin-like domain-CUT repeat-like tandem of special AT-rich sequence binding protein 1 (SATB1) reveals a coordinating DNA-binding mechanism.
J.Biol.Chem., 289, 2014
|
|
6EO6
 
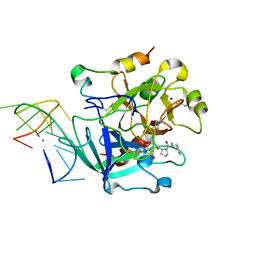 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(2-(1H-indol-3-yl)acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA63A - TBA MODIFIED APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
6EO7
 
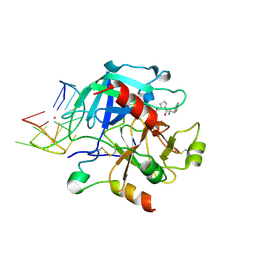 | | X-ray structure of the complex between human alpha-thrombin and modified 15-mer DNA aptamer containing 5-(3-(acetamide-N-yl)-1-propen-1-yl)-2'-deoxyuridine residue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GA68B2 - MODIFIED HUMAN THROMBIN BINDING APTAMER, ... | | Authors: | Dolot, R.M, Nawrot, B, Yang, X. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity.
Nucleic Acids Res., 46, 2018
|
|
3FGQ
 
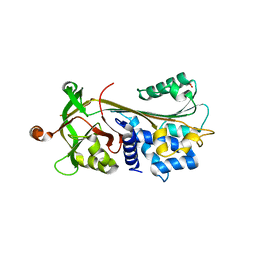 | | Crystal structure of native human neuroserpin | | Descriptor: | GLYCEROL, Neuroserpin | | Authors: | Takehara, S, Yang, X, Mikami, B, Onda, M. | | Deposit date: | 2008-12-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The 2.1-A crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability
J.Mol.Biol., 388, 2009
|
|
1R5K
 
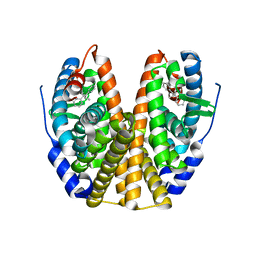 | | Human Estrogen Receptor alpha Ligand-Binding Domain In Complex With GW5638 | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, Estrogen receptor | | Authors: | Wu, Y.-L, Yang, X, Ren, Z, McDonnell, D.P, Norris, J.D, Willson, T.M, Greene, G.L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for an unexpected mode of SERM-mediated ER antagonism.
Mol.Cell, 18, 2005
|
|
1RKT
 
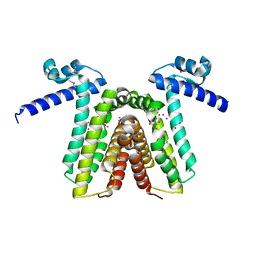 | | Crystal structure of yfiR, a putative transcriptional regulator from Bacillus subtilis | | Descriptor: | UNKNOWN ATOM OR ION, protein yfiR | | Authors: | Anderson, W.F, Rajan, S.S, Yang, X, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-23 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of YfiR, an unusual TetR/CamR-type putative transcriptional regulator from Bacillus subtilis.
Proteins, 65, 2006
|
|
6PH2
 
 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH4
 
 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PH3
 
 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6PPS
 
 | | A blue light illuminated LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Fernandez, I, Shin, H, Gunawardana, S, Otero, L.H, Cerutti, M.L, Yang, X, Klinke, S, Goldbaum, F.A. | | Deposit date: | 2019-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
4R3Q
 
 | | Crystal structure of SYCE3 | | Descriptor: | Synaptonemal complex central element protein 3 | | Authors: | Lu, J, Feng, J, Zhou, W, Yang, X, Shen, Y. | | Deposit date: | 2014-08-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural insight into the central element assembly of the synaptonemal complex
Sci Rep, 4, 2014
|
|
4PO5
 
 | | Crystal structure of allophycocyanin B from Synechocystis PCC 6803 | | Descriptor: | Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, PHYCOCYANOBILIN, ... | | Authors: | Pang, P.P, Dong, L.L, Sun, Y.F, Zeng, X.L, Ding, W.L, Scheer, H, Yang, X, Zhao, K.H. | | Deposit date: | 2014-02-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
