5DDQ
 
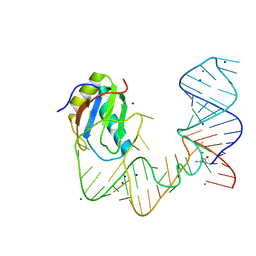 | | L-glutamine riboswitch bound with L-glutamine soaked with Mn2+ | | Descriptor: | GLUTAMINE, L-glutamine riboswitch RNA (61-MER), MAGNESIUM ION, ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
1FJB
 
 | | NMR Study of an 11-Mer DNA Duplex Containing 7,8-Dihydro-8-Oxoadenine (AOXO) Opposite Thymine | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(A38)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Chen, H, Johnson, F, Grollman, A.P, Patel, D.J. | | Deposit date: | 1995-12-15 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of the Ionizing Radiation Adduct 7,8-Dihydro-8-Oxoadenine (A Oxo) Positioned Opposite Thymine and Guanine in DNA Duplexes
MAGN.RESON.CHEM., 34, 1996
|
|
1D6D
 
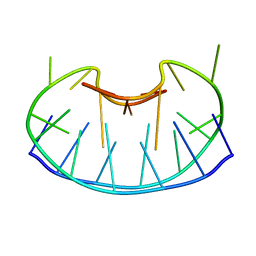 | | SOLUTION DNA STRUCTURE CONTAINING (A-A)-T TRIADS INTERDIGITATED BETWEEN A-T BASE PAIRS AND GGGG TETRADS; NMR, 8 STRUCT. | | Descriptor: | 5'-D(*AP*AP*GP*GP*TP*TP*TP*TP*AP*AP*GP*G)-3' | | Authors: | Kuryavyi, V.V, Kettani, A, Wang, W, Jones, R, Patel, D.J. | | Deposit date: | 1999-10-13 | | Release date: | 2000-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A diamond-shaped zipper-like DNA architecture containing triads sandwiched between mismatches and tetrads.
J.Mol.Biol., 295, 2000
|
|
5BNX
 
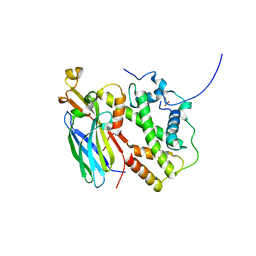 | |
1C0Y
 
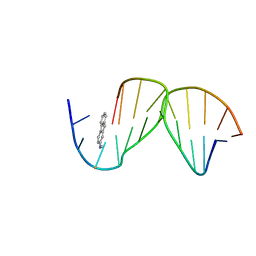 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED OPPOSITE DA AT A TEMPLATE-PRIMER JUNCTION | | Descriptor: | 2-AMINOFLUORENE, DNA (5'-D(*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Gu, Z, Gorin, A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1999-07-19 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction.
Biochemistry, 38, 1999
|
|
1FMN
 
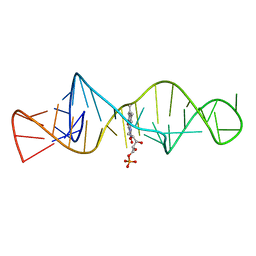 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | Authors: | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | Deposit date: | 1995-12-04 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
1FJA
 
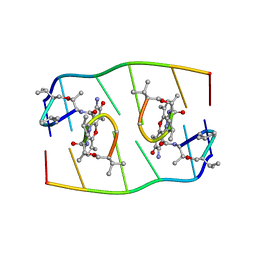 | |
3GII
 
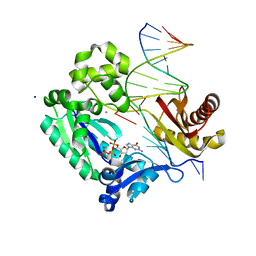 | | Dpo4 extension ternary complex with disordered A opposite an oxoG in anti conformation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(2DA))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
3GIL
 
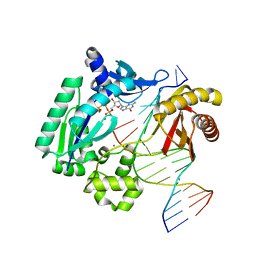 | | Dpo4 extension ternary complex with oxoG(anti)-T(anti) pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(2DT))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
5BS7
 
 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BNV
 
 | |
3GL6
 
 | |
1F3S
 
 | | Solution Structure of DNA Sequence GGGTTCAGG Forms GGGG Tetrade and G(C-A) Triad. | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*CP*AP*GP*G)-3') | | Authors: | Kettani, A, Basu, G, Gorin, A, Majumdar, A, Skripkin, E, Patel, D.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
J.Mol.Biol., 301, 2000
|
|
1G70
 
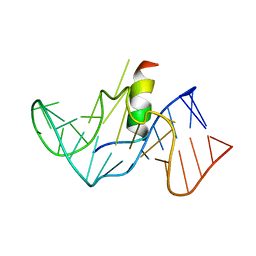 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
3DLH
 
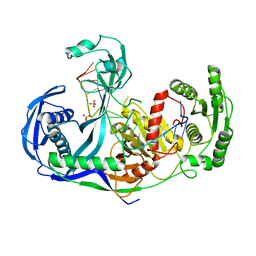 | | Crystal structure of the guide-strand-containing Argonaute protein silencing complex | | Descriptor: | ACETIC ACID, Argonaute, DNA (5'-D(DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DAP*DTP*DAP*DGP*DT)-3'), ... | | Authors: | Wang, Y, Sheng, G, Patel, D.J. | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the guide-strand-containing argonaute silencing complex.
Nature, 456, 2008
|
|
3DLB
 
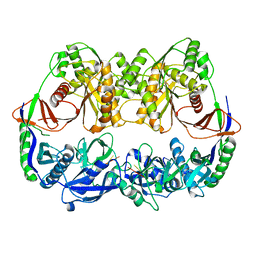 | |
3F73
 
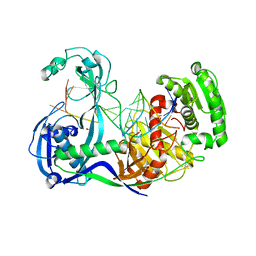 | | Alignment of guide-target seed duplex within an argonaute silencing complex | | Descriptor: | ARGONAUTE, DNA (5'-D(P*DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DA*DTP*DAP*DGP*DT)-3'), MAGNESIUM ION, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Juranek, S, Tuschl, T, Patel, D.J. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex.
Nature, 456, 2008
|
|
3O35
 
 | |
3O7X
 
 | |
3NNH
 
 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3OWW
 
 | |
3OXJ
 
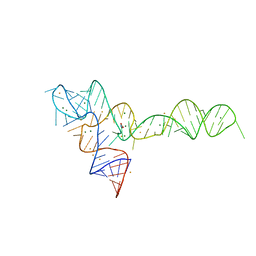 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | Descriptor: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3P04
 
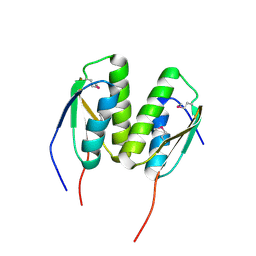 | | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8 | | Descriptor: | Uncharacterized BCR | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Hamilton, H, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8.
To be Published
|
|
6MUR
 
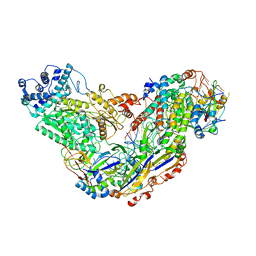 | | Cryo-EM structure of Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (27-MER), RNA (5'-R(P*GP*CP*AP*AP*AP*CP*CP*GP*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*C)-3'), Uncharacterized protein, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6MJW
 
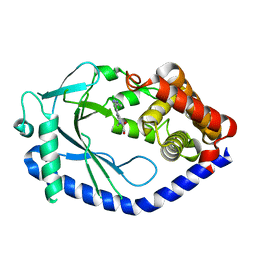 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
