4WZ3
 
 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
6AOJ
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal yeast Hog1p sequence | | Descriptor: | CHLORIDE ION, Ceg4, MAGNESIUM ION | | Authors: | Stogios, P.J, Nocek, B, Cuff, M.E, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
6BB9
 
 | | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-4-deoxychorismate lyase, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-17 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The crystal structure of 4-amino-4-deoxychorismate lyase from Salmonella typhimurium LT2
To Be Published
|
|
6BBX
 
 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
2R4Q
 
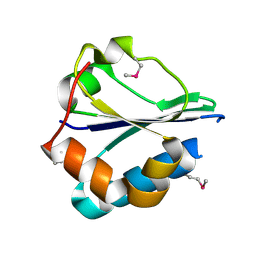 | | The structure of a domain of fruA from Bacillus subtilis | | Descriptor: | Phosphotransferase system (PTS) fructose-specific enzyme IIABC component | | Authors: | Cuff, M.E, Sather, A, Nocek, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a domain of fruA from Bacillus subtilis.
TO BE PUBLISHED
|
|
2R5R
 
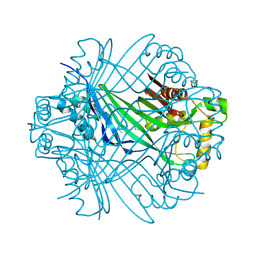 | | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718 | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, UPF0343 protein NE1163 | | Authors: | Tan, K, Wu, R, Nocek, B, Bigelow, L, Patterson, S, Freeman, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The crystal structure of DUF198 from Nitrosomonas europaea ATCC 19718.
To be Published
|
|
5UMY
 
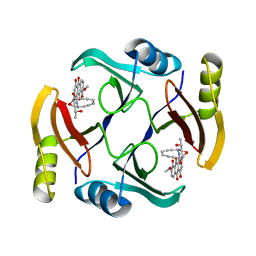 | | Crystal structure of TnmS3 in complex with tiancimycin | | Descriptor: | (1aS,11S,11aR,14Z,18R)-3,8,18-trihydroxy-11a-[(1R)-1-hydroxyethyl]-7-methoxy-11,11a-dihydro-4H-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinoline-4,9(10H)-dione, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMP
 
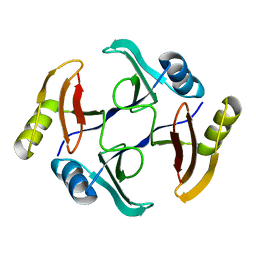 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
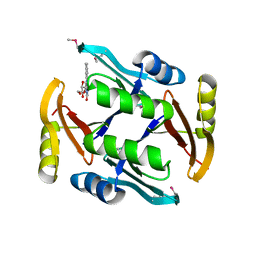 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
3V4Z
 
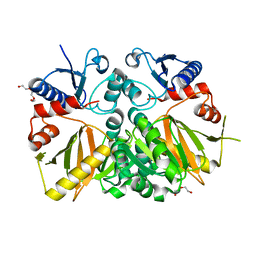 | | D-alanine--D-alanine ligase from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Nocek, B, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | D-alanine--D-alanine ligase from Yersinia pestis.
To be Published
|
|
5UMQ
 
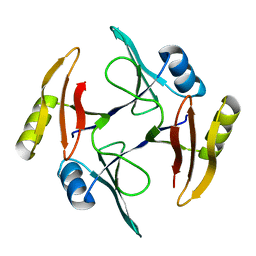 | | Crystal structure of TnmS1, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMX
 
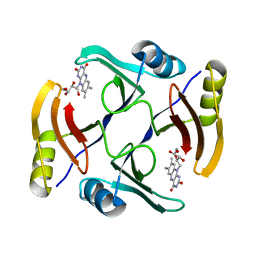 | | Crystal structure of TnmS3 in complex with riboflavin | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
3UZR
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside phosphotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form
TO BE PUBLISHED
|
|
5W27
 
 | | Crystal structure of TnmS3 in complex with tiancimycin (TNM B) | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-06-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
To Be Published
|
|
6B6L
 
 | | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Glycosyl hydrolase family 2, ... | | Authors: | Tan, K, Joachimiak, G, Nocek, B, Enddres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
6AOK
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal TEV protease cleavage sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ceg4, ... | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
6B7J
 
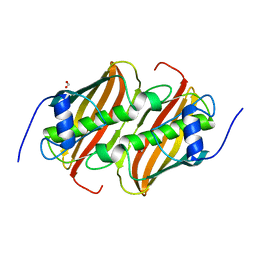 | |
6BMA
 
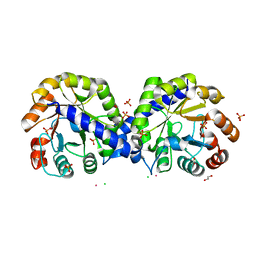 | | The crystal structure of indole-3-glycerol phosphate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Zhou, M, Nocek, B, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-14 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of indole-3-glycerol phosphate synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
4IYL
 
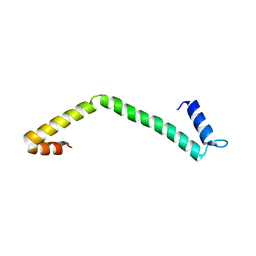 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
2C3C
 
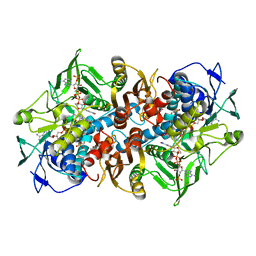 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2C3D
 
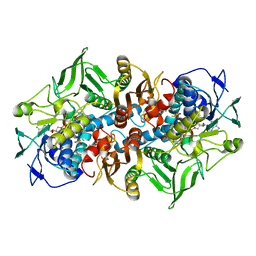 | | 2.15 Angstrom crystal structure of 2-ketopropyl coenzyme M oxidoreductase carboxylase with a coenzyme M disulfide bound at the active site | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
2ESH
 
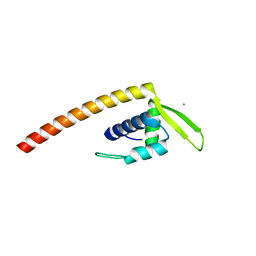 | | Crystal Structure of Conserved Protein of Unknown Function TM0937- a Potential Transcriptional Factor | | Descriptor: | CALCIUM ION, conserved hypothetical protein TM0937 | | Authors: | Liu, Y, Bochkareva, E, Zheng, H, Xu, X, Nocek, B, Lunin, V, Edward, A, Pai, E.F, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein TM0937
To be Published
|
|
4E16
 
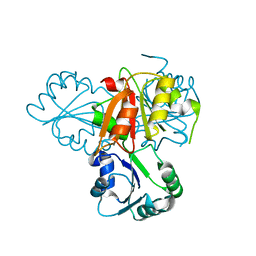 | | Precorrin-4 C(11)-methyltransferase from Clostridium difficile | | Descriptor: | precorrin-4 C(11)-methyltransferase | | Authors: | Osipiuk, J, Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-05 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Precorrin-4 C(11)-methyltransferase from Clostridium difficile
To be Published
|
|
4F3U
 
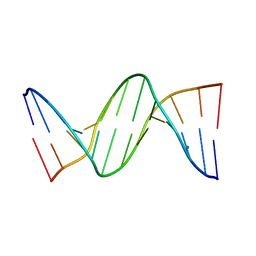 | |
3I0U
 
 | | Structure of the type III effector/phosphothreonine lyase OspF from Shigella flexneri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphothreonine lyase ospF | | Authors: | Singer, A.U, Skarina, T, Nocek, B, Gordon, R, Lam, R, Kagan, O, Edwards, A.M, Joachimiak, A, Chirgadze, N.Y, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the type III effector/phosphothreonine lyase OspF from Shigella flexneri
TO BE PUBLISHED
|
|
