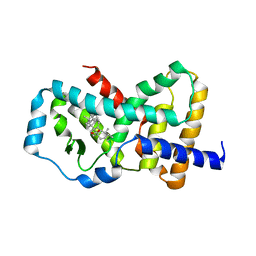
5K3M
 
 | |
6U26
 
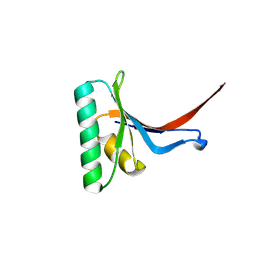 | | PCSK9 in complex with compound 16 | | Descriptor: | 4'-{[(1R)-6-{2-[2-({N~5~-[N,N'-bis(tert-butoxycarbonyl)carbamimidoyl]-N~2~-(tert-butoxycarbonyl)-L-ornithyl}amino)ethoxy]ethoxy}-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}-2'-fluoro[1,1'-biphenyl]-4-carboxylic acid, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Orth, P. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
6UCA
 
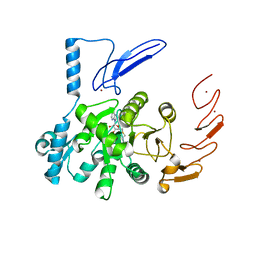 | | Crystal structure of human ZCCHC4 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, rRNA N6-adenosine-methyltransferase ZCCHC4 | | Authors: | Lu, J.W, Ren, W.D, Huang, M.J, Gao, L, Li, D.X, Wang, G.G, Song, J. | | Deposit date: | 2019-09-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA.
Nat Commun, 10, 2019
|
|
1QSQ
 
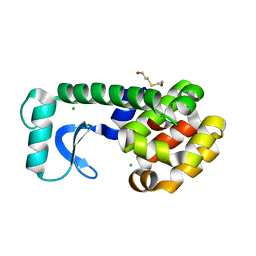 | | CAVITY CREATING MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Matthews, B.W. | | Deposit date: | 1999-06-22 | | Release date: | 1999-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
8E3X
 
 | | Cryo-EM structure of the PAC1R-PACAP27-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
8E3Z
 
 | | Cryo-EM structure of the VPAC1R-VIP-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
8E3Y
 
 | | Cryo-EM structure of the VPAC1R-PACAP27-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
8D56
 
 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
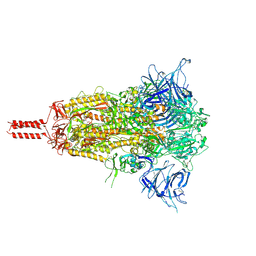 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D5A
 
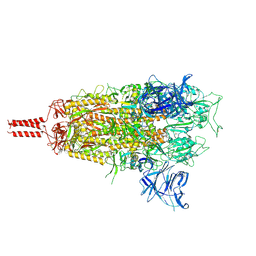 | | Middle state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6U8X
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpApG DNA | | Descriptor: | CpApG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95063877 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8P
 
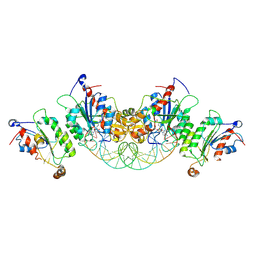 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpA DNA | | Descriptor: | CpGpA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-05 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
7KJ4
 
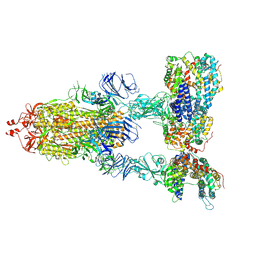 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ3
 
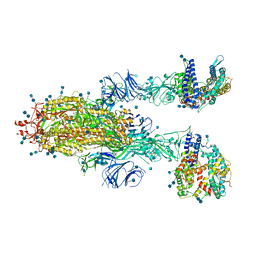 | | SARS-CoV-2 Spike Glycoprotein with two ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ5
 
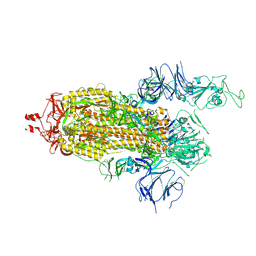 | | SARS-CoV-2 Spike Glycoprotein, prefusion with one RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ2
 
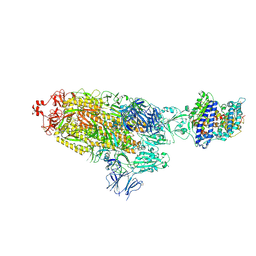 | | SARS-CoV-2 Spike Glycoprotein with one ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LCR
 
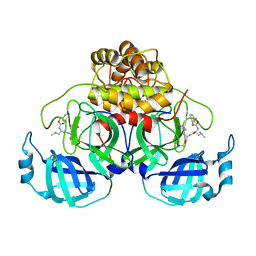 | | Improved Feline Drugs as SARS-CoV-2 Mpro Inhibitors: Structure-Activity Studies & Micellar Solubilization for Enhanced Bioavailability | | Descriptor: | 3C-like proteinase, N~2~-{[(3-fluorophenyl)methoxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Improved SARS-CoV-2 M pro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies.
Eur.J.Med.Chem., 222, 2021
|
|
7N1W
 
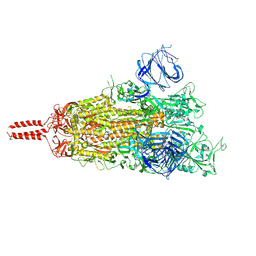 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1Y
 
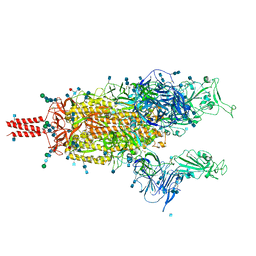 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1U
 
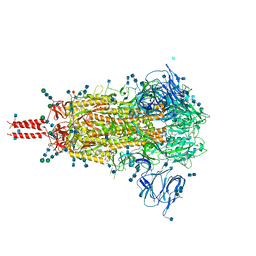 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1X
 
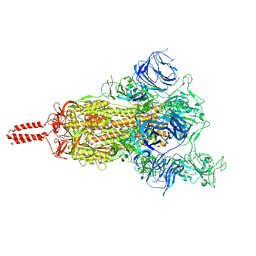 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1T
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1Q
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1V
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7XCZ
 
 | |
