2JRX
 
 | | Solution NMR structure of protein YejL from E. coli. Northeast Structural Genomics target ER309 | | Descriptor: | UPF0352 protein yejL | | Authors: | Cort, J.R, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Chemical Shift Assignments of E. coli YejL protein.
To be Published
|
|
2JOE
 
 | | NMR Structure of E. Coli YehR Protein. Northeast Structural Genomics Target ER538. | | Descriptor: | Hypothetical lipoprotein yehR | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of E. Coli YehR Protein
To be Published
|
|
2K2E
 
 | | Solution NMR structure of Bordetella pertussis protein BP2786, a Mth938-like domain. Northeast Structural Genomics Consortium target BeR31 | | Descriptor: | Uncharacterized protein BP2786 | | Authors: | Cort, J.R, Ho, C.K, Nwosu, C, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bordetella pertussis protein BP2786, a Mth938-like domain.
To be Published
|
|
2K31
 
 | | Solution Structure of cGMP-binding GAF domain of Phosphodiesterase 5 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, Phosphodiesterase 5A, cGMP-specific | | Authors: | Heikaus, C.C, Stout, J.R, Sekharan, M.R, Eakin, C.M, Rajagopal, P, Brzovic, P.S, Beavo, J.A, Klevit, R.E. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the cGMP Binding GAF Domain from Phosphodiesterase 5: Insights into Nucleotide Selectivity, Dimerization, and cGMP-Dependent Conformational Change.
J.Biol.Chem., 283, 2008
|
|
2JUZ
 
 | | Solution NMR structure of HI0947 from Haemophilus influenzae, Northeast Structural Genomics Consortium Target IR123 | | Descriptor: | UPF0352 protein HI0840 | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Wang, D, Nwosu, C, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-09 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of HI0947 from Haemophilus influenzae.
To be Published
|
|
2LFM
 
 | |
2L77
 
 | |
2LEK
 
 | | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris refined with NH RDCs. Northeast Structural Genomics Consortium target RpR325 | | Descriptor: | Putative thiamin biosynthesis ThiS | | Authors: | Ramelot, T.A, Cort, J.R, Lee, H, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium target RpR325
To be Published
|
|
2L8V
 
 | | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC (20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR219A | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC
(20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics
Consortium Target TeR219A
To be Published
|
|
2LAK
 
 | | Solution NMR structure of the AHSA1-like protein RHE_CH02687 (1-152) from Rhizobium etli, Northeast Structural Genomics Consortium Target ReR242 | | Descriptor: | AHSA1-like protein RHE_CH02687 | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-16 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein RHE_CH02687 (1-152) from Rhizobium etli, Northeast Structural Genomics Consortium Target ReR242
To be Published
|
|
2L06
 
 | | Solution NMR structure of the PBS linker polypeptide domain (fragment 254-400) of phycobilisome linker protein ApcE from Synechocystis sp. PCC 6803. Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | Phycobilisome LCM core-membrane linker polypeptide | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Hamilton, K, Ciccosanti, C, Lee, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the PBS linker polypeptide domain of phycobilisome
linker protein apcE from Synechocystis sp. Northeast Structural Genomics Consortium
Target SgR209C
To be Published
|
|
2LOA
 
 | | Structural Basis for Bifunctional Zn(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA | | Descriptor: | 4-(1,4,7,10-tetraazacyclododecan-1-ylmethyl)quinoline, DNA (5'-D(*GP*CP*CP*GP*CP*AP*GP*TP*GP*C)-3'), ZINC ION | | Authors: | Morrow, J.R, Fountain, M.A, del Mundo, I.A, Siter, K.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Bifunctional Zinc(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA.
Inorg.Chem., 51, 2012
|
|
2LP6
 
 | | Refined Solution NMR Structure of the 50S ribosomal protein L35Ae from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target (NESG) PfR48 | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Yu, B, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-02 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ribosomal protein RP-L35Ae from Pyrococcus furiosus.
Proteins, 80, 2012
|
|
2LO8
 
 | |
2LO5
 
 | | Structural Basis for Bifunctional Zn(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA, Structure of a Thymine bulge | | Descriptor: | DNA (5'-D(*GP*GP*CP*CP*GP*CP*AP*GP*TP*GP*CP*C)-3') | | Authors: | Morrow, J.R, Fountain, M.A, del Mundo, I.A, Siter, K.E. | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Bifunctional Zinc(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA.
Inorg.Chem., 51, 2012
|
|
1SD1
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH FORMYCIN A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine phosphorylase | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
1SD2
 
 | | STRUCTURE OF HUMAN 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH 5'-METHYLTHIOTUBERCIDIN | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 5'-methylthioadenosine phosphorylase, SULFATE ION | | Authors: | Lee, J.E, Settembre, E.C, Cornell, K.A, Riscoe, M.K, Sufrin, J.R, Ealick, S.E, Howell, P.L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Comparison of MTA Phosphorylase and MTA/AdoHcy Nucleosidase Explains Substrate Preferences and Identifies Regions Exploitable for Inhibitor Design.
Biochemistry, 43, 2004
|
|
1SOU
 
 | | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46 | | Descriptor: | 5,10-methenyltetrahydrofolate synthetase | | Authors: | Cort, J.R, Chiang, Y, Acton, T, Wu, M, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Aquifex aeolicus 5,10-methenyltetrahydrofolate synthetase: Northeast Structural Genomics Consortium Target QR46
To be Published
|
|
1UD7
 
 | | SOLUTION STRUCTURE OF THE DESIGNED HYDROPHOBIC CORE MUTANT OF UBIQUITIN, 1D7 | | Descriptor: | PROTEIN (UBIQUITIN CORE MUTANT 1D7) | | Authors: | Johnson, E.C, Lazar, G.A, Desjarlais, J.R, Handel, T.M. | | Deposit date: | 1999-04-07 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin.
Structure Fold.Des., 7, 1999
|
|
6MG1
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta, ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
1U35
 
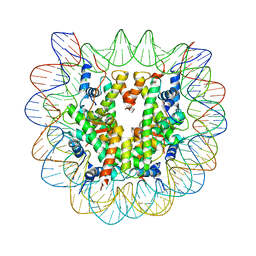 | | Crystal structure of the nucleosome core particle containing the histone domain of macroH2A | | Descriptor: | H2A histone family, Hist1h4i protein, Histone H3.1, ... | | Authors: | Chakravarthy, S, Gundimella, S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of the histone variant macroH2A.
Mol.Cell.Biol., 25, 2005
|
|
1S98
 
 | | E.coli IscA crystal structure to 2.3 A | | Descriptor: | Protein yfhF | | Authors: | Cupp-Vickery, J.R, Silberg, J.J, Ta, D.T, Vickery, L.E. | | Deposit date: | 2004-02-03 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IscA, an iron-sulfur cluster assembly protein from Escherichia coli.
J.Mol.Biol., 338, 2004
|
|
6MBK
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, First P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
1S3Y
 
 | | Structure Determination of Tetrahydroquinazoline Antifolates in Complex with Human and Pneumocystis carinii Dihydrofolate Reductase: Correlations of Enzyme Selectivity and Stereochemistry | | Descriptor: | 6-(OCTAHYDRO-1H-INDOL-1-YLMETHYL)DECAHYDROQUINAZOLINE-2,4-DIAMINE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A, Queener, S.F. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure determination of tetrahydroquinazoline antifolates in complex with human and Pneumocystis carinii dihydrofolate reductase: correlations between enzyme selectivity and stereochemistry.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6MBO
 
 | | GLP Methyltransferase with Inhibitor EML741-P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
