5YNR
 
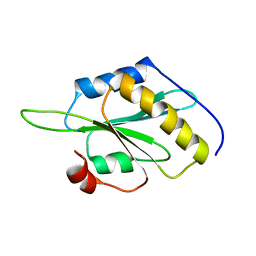 | |
4XCI
 
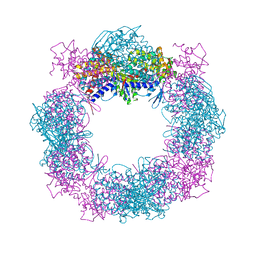 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form II | | Descriptor: | Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Chaston, J.J, Smits, C, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
6MH6
 
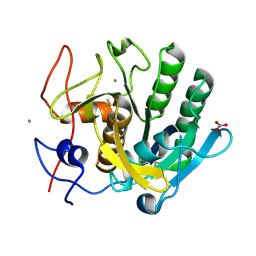 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6FR2
 
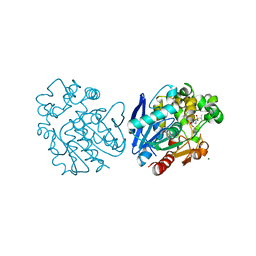 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
8UTK
 
 | | IL-23R minibinder - 23R-B04dslf02IB | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 23R-B04dslf02IB, ... | | Authors: | Bera, A.K, Berger, S.A, Kang, A, Baker, D. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Preclinical proof of principle for orally delivered Th17 antagonist miniproteins.
Cell, 187, 2024
|
|
8JSX
 
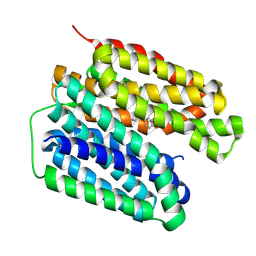 | | VMAT2 complex with noradrenaline in lumen-facing state | | Descriptor: | Noradrenaline, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-20 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8JTB
 
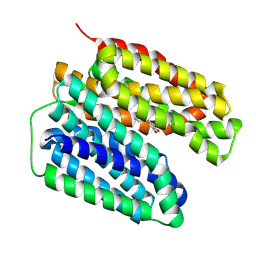 | | VMAT2 complex with dopamine | | Descriptor: | L-DOPAMINE, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
8JT5
 
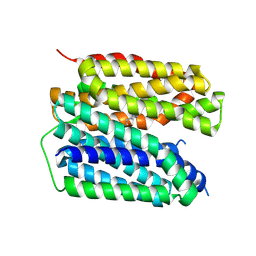 | | VMAT2 complex with histamine | | Descriptor: | HISTAMINE, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
3AG9
 
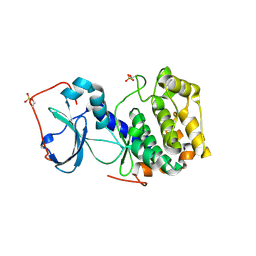 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 | | Descriptor: | (10R,20R,23R)-10-(4-aminobutyl)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamidopropyl)-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Lavogina, D, Uri, A, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2010-03-26 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8USN
 
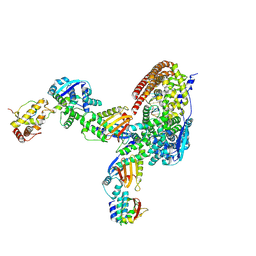 | | Intracellular cryo-tomography structure of EBOV nucleocapsid at 8.9 Angstrom | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein, Polymerase cofactor VP35, ... | | Authors: | Watanabe, R, Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.
Cell, 187, 2024
|
|
8UST
 
 | | In-virion structure of Ebola virus nucleocapsid-like assemblies from recombinant virus-like particles (nucleoprotein, VP24,VP35,VP40) | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein, Polymerase cofactor VP35, ... | | Authors: | Watanabe, R, Zyla, D, Saphire, E.O. | | Deposit date: | 2023-10-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Intracellular Ebola virus nucleocapsid assembly revealed by in situ cryo-electron tomography.
Cell, 187, 2024
|
|
2Y68
 
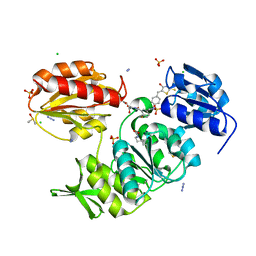 | | Structure-based design of a new series of D-glutamic acid-based inhibitors of bacterial MurD ligase | | Descriptor: | 2-[[2-fluoro-5-[[[4-[(Z)-(4-oxo-2-sulfanylidene-1,3-thiazolidin-5-ylidene)methyl]phenyl]amino]methyl]phenyl]carbonylamino]pentanedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-based design of a new series of D-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD).
J. Med. Chem., 54, 2011
|
|
6MH8
 
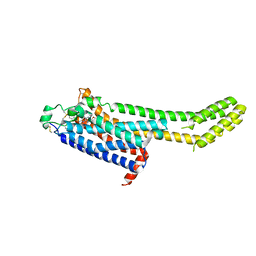 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
1HCP
 
 | |
2YCT
 
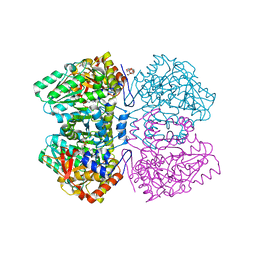 | | Tyrosine phenol-lyase from Citrobacter freundii in complex with pyridine N-oxide and the quinonoid intermediate formed with L-alanine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}PROPANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PHOSPHATE ION, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
2Y8L
 
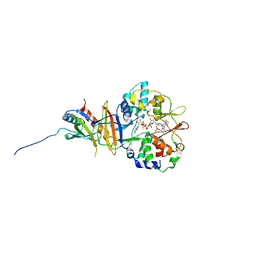 | | Structure of the regulatory fragment of mammalian aMPK in complex with two ADP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
7TAS
 
 | |
2YA3
 
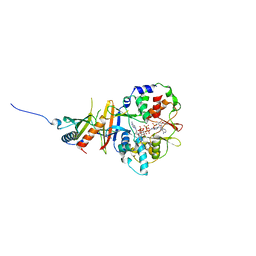 | | STRUCTURE OF THE REGULATORY FRAGMENT OF MAMMALIAN AMPK IN COMPLEX WITH COUMARIN ADP | | Descriptor: | 3'-(7-diethylaminocoumarin-3-carbonylamino)-3'-deoxy-ADP, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
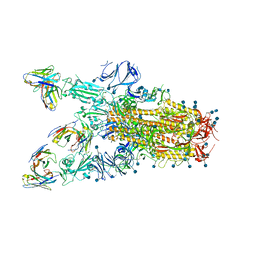
7TAT
 
 | |
2YHK
 
 | | D214A mutant of tyrosine phenol-lyase from Citrobacter freundii | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, DI(HYDROXYETHYL)ETHER, POTASSIUM ION, ... | | Authors: | Milic, D, Demidkina, T.V, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-05-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Citrobacter Freundii Asp214Ala Tyrosine Phenol-Lyase Reveals that Asp214 is Critical for Maintaining a Strain in the Internal Aldimine
Croatica Chemica Acta, 85, 2012
|
|
2YFD
 
 | | STRUCTURAL AND FUNCTIONAL INSIGHTS OF DR2231 PROTEIN, THE MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE FROM DEINOCOCCUS RADIODURANS, COMPLEXED WITH Mg and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goncalves, A.M.D, De Sanctis, D, Mcsweeney, S.M. | | Deposit date: | 2011-04-05 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
6XYE
 
 | |
8VLW
 
 | |
6IOZ
 
 | | Structural insights of idursulfase beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, H, Kim, D, Hong, J, Lee, K, Seo, J, Oh, B.H. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights of idursulfase beta
To Be Published
|
|
