3M8U
 
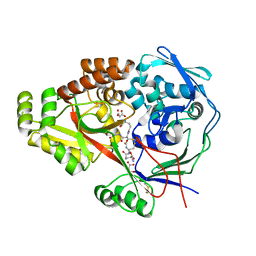 | | Crystal structure of glutathione-binding protein A (GbpA) from Haemophilus parasuis SH0165 in complex with glutathione disulfide (GSSG) | | Descriptor: | Heme-binding protein A, MALONATE ION, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Vergauwen, B, Elegheert, J, Dansercoer, A, Devreese, B, Savvides, S.N. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6JWZ
 
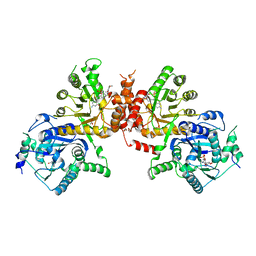 | | Crystal structure of Plasmodium falciparum HPPK-DHPS S436F/A437G/A613S triple mutant with SDX-DHP | | Descriptor: | 4-[(2-azanyl-4-oxidanylidene-7,8-dihydro-3~{H}-pteridin-6-yl)methylamino]-~{N}-(5,6-dimethoxypyrimidin-4-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ACETATE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
5UXB
 
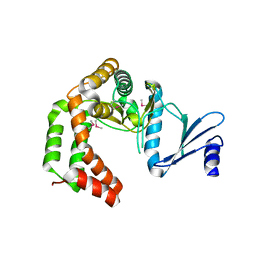 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
7EQM
 
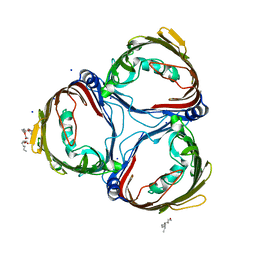 | | Apo Truncated VhChiP (Delta 1-19) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, MAGNESIUM ION, ... | | Authors: | Aunkham, A, Sanram, S, Suginta, W. | | Deposit date: | 2021-05-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.50002384 Å) | | Cite: | Structural displacement model of chitooligosaccharide transport through chitoporin.
J.Biol.Chem., 299, 2023
|
|
6CAA
 
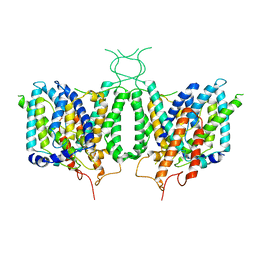 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
6C36
 
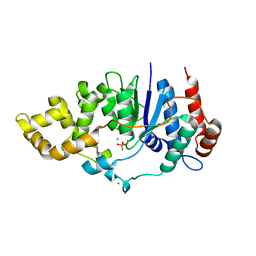 | | Mycobacterium smegmatis flap endonuclease mutant D208N | | Descriptor: | 5'-3' exonuclease, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Shuman, S, Goldgur, Y, Carl, A, Uson, M.L. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Nucleic Acids Res., 46, 2018
|
|
7FD2
 
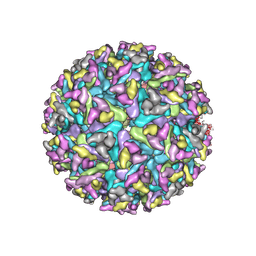 | | Cryo-EM structure of an alphavirus, Getah virus | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, Z, Liu, C, Wang, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of infective Getah virus at 2.8 angstrom resolution determined by cryo-electron microscopy.
Cell Discov, 8, 2022
|
|
7EQR
 
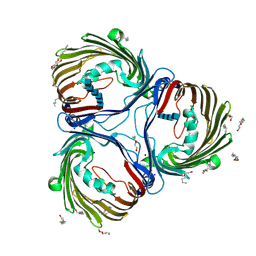 | | Crystal structure of Truncated (Delta 1-19) Chitoporin VhChiP from Vibrio harveyi in complex with chitohexaose | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aunkham, A, Sanram, S, Suginta, S. | | Deposit date: | 2021-05-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.75003171 Å) | | Cite: | Structural displacement model of chitooligosaccharide transport through chitoporin.
J.Biol.Chem., 299, 2023
|
|
5V0S
 
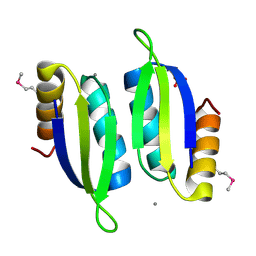 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | Descriptor: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6K24
 
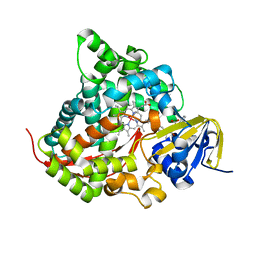 | | Structure of the Rhodium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7EXH
 
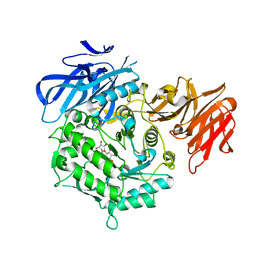 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactinol. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, galactinol | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXR
 
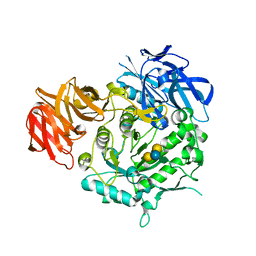 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with Stachyose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXF
 
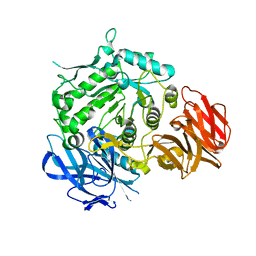 | | Crystal structure of wild-type from Arabidopsis thaliana complexed with Galactose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5V6P
 
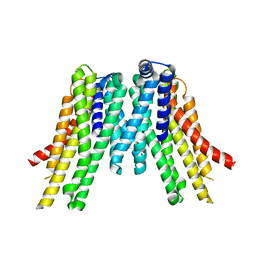 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
7EXJ
 
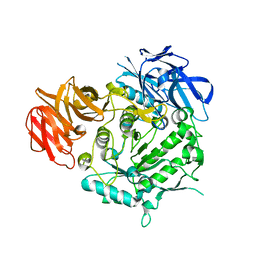 | | Crystal structure of alkaline alpha-galctosidase D383A mutant from Arabidopsis thaliana complexed with Raffinose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXG
 
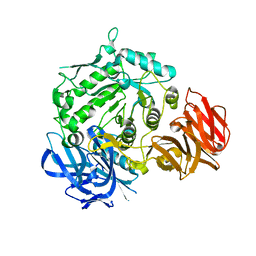 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7F96
 
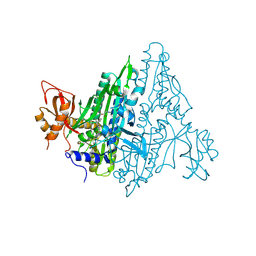 | | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with L-proline and compound L95 | | Descriptor: | PROLINE, Proline--tRNA ligase, ~{N}-[4-[(3~{S})-3-cyano-3-cyclopropyl-2-oxidanylidene-pyrrolidin-1-yl]-6-methyl-pyridin-2-yl]-2-phenyl-ethanamide | | Authors: | Mishra, S, Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-07-04 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | ATP mimetics targeting prolyl-tRNA synthetases as a new avenue for antimalarial drug development
Iscience, 27, 2024
|
|
7EXQ
 
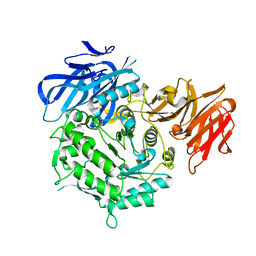 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with product-galactose and sucrose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7F97
 
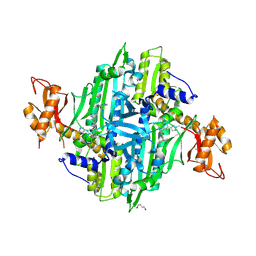 | | Plasmodium falciparum Prolyl-tRNA Synthetase (PfPRS) in Complex with L-proline and compound L97 | | Descriptor: | 1,4-BUTANEDIOL, 4-[(3S)-3-cyclopropyl-3-(hydroxymethyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, CHLORIDE ION, ... | | Authors: | Mishra, S, Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-07-04 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | ATP mimetics targeting prolyl-tRNA synthetases as a new avenue for antimalarial drug development
Iscience, 27, 2024
|
|
5UQ4
 
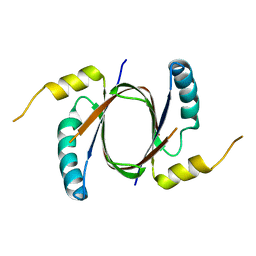 | | Crystal structure of Heme-Degrading Protein Rv3592 from Mycobacterium tuberculosis - heme free with cleaved protein | | Descriptor: | Monooxygenase | | Authors: | Chang, C, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-06 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal structure of Heme-Degrading Protein Rv3592 from Mycobacterium tuberculosis - heme free with cleaved protein.
To Be Published
|
|
4GRW
 
 | | Structure of a complex of human IL-23 with 3 Nanobodies (Llama vHHs) | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, Nanobody 124C4, ... | | Authors: | Desmyter, A, Spinelli, S, Button, C, Saunders, M, de Haard, H, Rommelaere, H, Union, A, Cambillau, C. | | Deposit date: | 2012-08-27 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Neutralization of Human Interleukin 23 by Multivalent Nanobodies Explained by the Structure of Cytokine-Nanobody Complex.
Front Immunol, 8, 2017
|
|
3JYO
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Niefind, K, Schomburg, D. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6J61
 
 | | Crystal Structure of Thymidylate Synthase, Thy1, from Thermus thermophilus having an Extra C Terminal Domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, PHOSPHATE ION | | Authors: | Ogawa, A, Sampei, G, Kawai, G. | | Deposit date: | 2019-01-12 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the flavin-dependent thymidylate synthase Thy1 from Thermus thermophilus with an extra C-terminal domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1G51
 
 | | ASPARTYL TRNA SYNTHETASE FROM THERMUS THERMOPHILUS AT 2.4 A RESOLUTION | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, ASPARTYL-TRNA SYNTHETASE, ... | | Authors: | Poterzsman, A, Delarue, M, Thierry, J.C, Moras, D. | | Deposit date: | 2000-10-30 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase.
J.Mol.Biol., 244, 1994
|
|
