7EWZ
 
 | |
7EX6
 
 | |
7EX0
 
 | |
7EX8
 
 | | Crystal structure of Ebinur Lake virus cap snatching endonuclease in complex with inhibitor 27 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Replicase, ... | | Authors: | Kuang, W, Hu, Z, Gong, P. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biochemical Basis for Development of Diketo Acid Inhibitors Targeting the Cap-Snatching Endonuclease of the Ebinur Lake Virus (Order: Bunyavirales ).
J.Virol., 96, 2022
|
|
7EX9
 
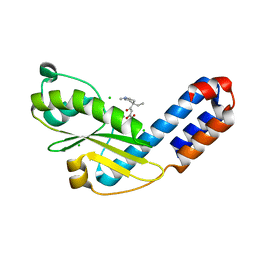 | | Crystal structure of Ebinur Lake virus cap snatching endonuclease in complex with inhibitor 28 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Replicase, ... | | Authors: | Kuang, W, Hu, Z, Gong, P. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Basis for Development of Diketo Acid Inhibitors Targeting the Cap-Snatching Endonuclease of the Ebinur Lake Virus (Order: Bunyavirales ).
J.Virol., 96, 2022
|
|
7EX2
 
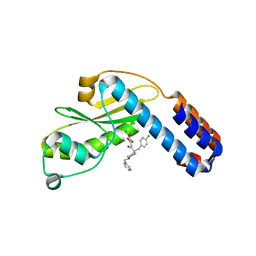 | | Crystal structure of Ebinur Lake virus cap snatching endonuclease in complex with L-742,001 (methyl ester form) | | Descriptor: | MANGANESE (II) ION, Replicase, methyl (Z)-4-[4-[(4-chlorophenyl)methyl]-1-(phenylmethyl)piperidin-4-yl]-2-oxidanyl-4-oxidanylidene-but-2-enoate | | Authors: | Kuang, W, Hu, Z, Gong, P. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Biochemical Basis for Development of Diketo Acid Inhibitors Targeting the Cap-Snatching Endonuclease of the Ebinur Lake Virus (Order: Bunyavirales ).
J.Virol., 96, 2022
|
|
7EX4
 
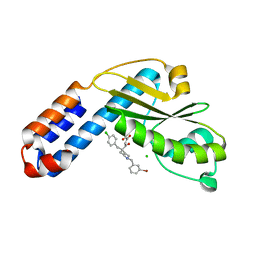 | | Crystal structure of Ebinur Lake virus cap snatching endonuclease in complex with inhibitor 5 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Replicase, ... | | Authors: | Kuang, W, Hu, Z, Gong, P. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Basis for Development of Diketo Acid Inhibitors Targeting the Cap-Snatching Endonuclease of the Ebinur Lake Virus (Order: Bunyavirales ).
J.Virol., 96, 2022
|
|
7EV3
 
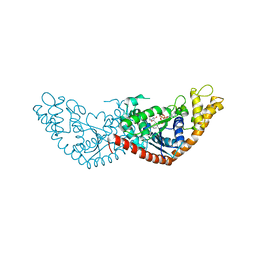 | |
7EX7
 
 | | Crystal structure of Ebinur Lake virus cap snatching endonuclease in complex with inhibitor 16 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Replicase, ... | | Authors: | Kuang, W, Hu, Z, Gong, P. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Basis for Development of Diketo Acid Inhibitors Targeting the Cap-Snatching Endonuclease of the Ebinur Lake Virus (Order: Bunyavirales ).
J.Virol., 96, 2022
|
|
7EX3
 
 | | Crystal structure of Ebinur Lake virus cap snatching endonuclease in complex with inhibitor 3 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Replicase, ... | | Authors: | Kuang, W, Hu, Z, Gong, P. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Biochemical Basis for Development of Diketo Acid Inhibitors Targeting the Cap-Snatching Endonuclease of the Ebinur Lake Virus (Order: Bunyavirales ).
J.Virol., 96, 2022
|
|
7EV2
 
 | |
7F45
 
 | | Structure of an Anti-CRISPR protein | | Descriptor: | AcrIF5 | | Authors: | Feng, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
7EQG
 
 | | Structure of Csy-AcrIF5 | | Descriptor: | AcrIF5, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | AcrIF5 specifically targets DNA-bound CRISPR-Cas surveillance complex for inhibition.
Nat.Chem.Biol., 18, 2022
|
|
5YAX
 
 | | Crystal structure of a human neutralizing antibody bound to a HBV preS1 peptide | | Descriptor: | Large envelope protein, SODIUM ION, scFv1 antibody | | Authors: | Liu, X, Zheng, S, Ye, K, Sui, J. | | Deposit date: | 2017-09-02 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Elife, 6, 2017
|
|
5C0M
 
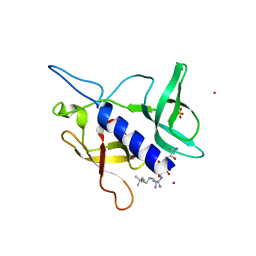 | | Crystal structure of SGF29 tandem tudor domain in complex with a Carba containing peptide | | Descriptor: | Carba-containing peptide, GLYCEROL, SAGA-associated factor 29 homolog, ... | | Authors: | Dong, A, Xu, C, Tempel, W, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical basis for the recognition of trimethyllysine by epigenetic reader proteins.
Nat Commun, 6, 2015
|
|
7LUA
 
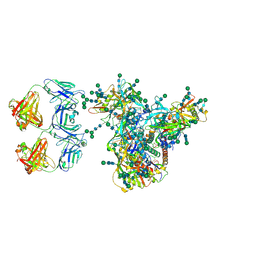 | | Cryo-EM structure of DH898.1 Fab-dimer bound near the CD4 binding site of HIV-1 Env CH848 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 SOSIP gp120, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-17 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
7LU9
 
 | | Cryo-EM structure of DH851.3 bound to HIV-1 CH505 Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Manne, K, Edwards, R.J, Acharya, P. | | Deposit date: | 2021-02-21 | | Release date: | 2021-03-24 | | Last modified: | 2021-06-09 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
4ZOL
 
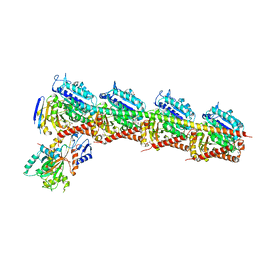 | | Crystal Structure of Tubulin-Stathmin-TTL-Tubulysin M Complex | | Descriptor: | (2R,4R)-4-{[(2-{(1R,3R)-1-(acetyloxy)-4-methyl-3-[methyl(N-{[(2S)-1-methylpiperidin-2-yl]carbonyl}-D-isoleucyl)amino]pentyl}-1,3-thiazol-4-yl)carbonyl]amino}-2-methyl-5-phenylpentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-05-06 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
1XC3
 
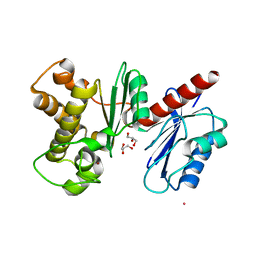 | | Structure of a Putative Fructokinase from Bacillus subtilis | | Descriptor: | GLYCEROL, PLATINUM (II) ION, Putative fructokinase, ... | | Authors: | Cuff, M.E, Quartey, P, Lezondra, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of ROK fructokinase YdhR from Bacillus subtilis: insights into substrate binding and fructose specificity.
J.Mol.Biol., 406, 2011
|
|
4ZKT
 
 | |
4Z7P
 
 | | X-ray structure of racemic ShK Q16K toxin | | Descriptor: | Potassium channel toxin kappa-stichotoxin-She1a, SULFATE ION | | Authors: | Sickmier, E.A. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-09 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Pharmaceutical Optimization of Peptide Toxins for Ion Channel Targets: Potent, Selective, and Long-Lived Antagonists of Kv1.3.
J.Med.Chem., 58, 2015
|
|
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
4Y7N
 
 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4ZHQ
 
 | | Crystal structure of Tubulin-Stathmin-TTL-MMAE Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
4ZI7
 
 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-HTI286 COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
