6ACK
 
 | |
8H6T
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6P
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
2ZNV
 
 | | Crystal structure of human AMSH-LP DUB domain in complex with Lys63-linked ubiquitin dimer | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, Ubiquitin, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
6ACC
 
 | |
6ACD
 
 | |
2ZNR
 
 | | Crystal structure of the DUB domain of human AMSH-LP | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, PRASEODYMIUM ION, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
8GYD
 
 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
6KQI
 
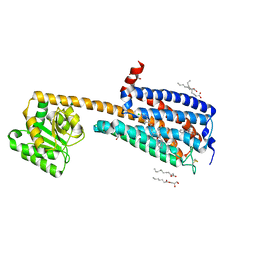 | | Structure of an allosteric modulator bound to the CB1 cannabinoid receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 5-chloro-3-ethyl-N-{2-[4-(piperidin-1-yl)phenyl]ethyl}-1H-indole-2-carboxamide, ... | | Authors: | Shao, Z.H, Yan, W. | | Deposit date: | 2019-08-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Structure of an allosteric modulator bound to the CB1 cannabinoid receptor.
Nat.Chem.Biol., 15, 2019
|
|
6LL9
 
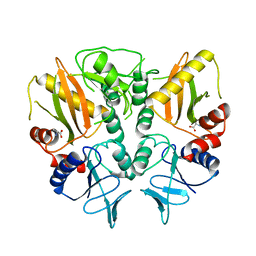 | |
6IQL
 
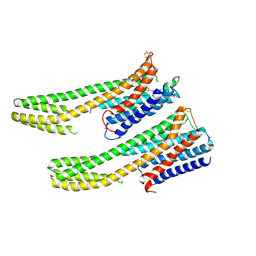 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | Descriptor: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | Authors: | Zhou, Y, Cao, C, Zhang, X.C. | | Deposit date: | 2018-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
6MH4
 
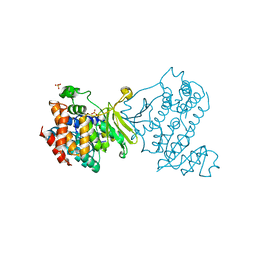 | |
6MH5
 
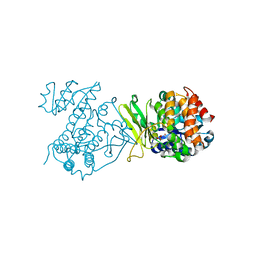 | |
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7WN1
 
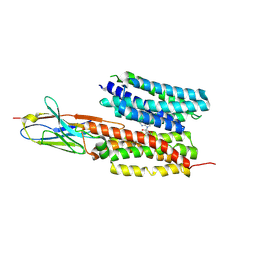 | | Structure of PfNT1(Y190A) in complex with nanobody 48 and inosine | | Descriptor: | Equilibrative nucleoside/nucleobase transporter, INOSINE, nanobody48 | | Authors: | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | Deposit date: | 2022-01-17 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
6JXA
 
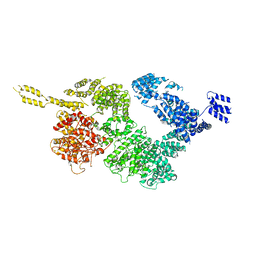 | | Tel1 kinase compact monomer | | Descriptor: | Serine/threonine-protein kinase TEL1 | | Authors: | Xin, J. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of allosteric regulation of Tel1/ATM kinase.
Cell Res., 29, 2019
|
|
7LIX
 
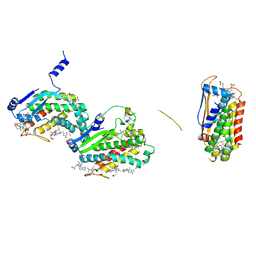 | | CaRSP1 and scaffolded phycoerythrin beta subunits from the phycobilisome of Porphyridium purpureum | | Descriptor: | B-phycoerythrin beta chain, CaRSP1, PHYCOERYTHROBILIN | | Authors: | Rathbone, H.W, Landsberg, M.J, Michie, K.A, Green, B.R, Curmi, P.M.G. | | Deposit date: | 2021-01-28 | | Release date: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Scaffolding proteins guide the evolution of algal light harvesting antennas.
Nat Commun, 12, 2021
|
|
7LJ0
 
 | | Linker 3 and scaffolded phycoerythrin beta subunit from the phycobilisome of Porphyridium purpureum | | Descriptor: | B-phycoerythrin beta chain, Linker 3, PHYCOERYTHROBILIN | | Authors: | Rathbone, H.W, Landsberg, M.J, Michie, K.A, Green, B.R, Curmi, P.M.G. | | Deposit date: | 2021-01-28 | | Release date: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Scaffolding proteins guide the evolution of algal light harvesting antennas.
Nat Commun, 12, 2021
|
|
7LIY
 
 | | CaRSP2 and scaffolded phycoerythrin beta subunits from the phycobilisome of Porphyridium purpureum | | Descriptor: | B-phycoerythrin beta chain, CaRSP2, PHYCOERYTHROBILIN | | Authors: | Rathbone, H.W, Landsberg, M.J, Michie, K.A, Green, B.R, Curmi, P.M.G. | | Deposit date: | 2021-01-28 | | Release date: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Scaffolding proteins guide the evolution of algal light harvesting antennas.
Nat Commun, 12, 2021
|
|
7LIZ
 
 | | LR6 rod linker and scaffolded phycoerythrin beta subunits from the phycobilisome of Porphyridium purpureum | | Descriptor: | B-phycoerythrin beta chain, LR6, PHYCOERYTHROBILIN | | Authors: | Rathbone, H.W, Landsberg, M.J, Michie, K.A, Green, B.R, Curmi, P.M.G. | | Deposit date: | 2021-01-28 | | Release date: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Scaffolding proteins guide the evolution of algal light harvesting antennas.
Nat Commun, 12, 2021
|
|
6JSH
 
 | | Apo-state Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Qiu, S.W, Liu, S. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
6JSI
 
 | | Co-purified Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Qiu, S.W, Liu, S. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
7WH8
 
 | |
6JXC
 
 | | Tel1 kinase butterfly symmetric dimer | | Descriptor: | Serine/threonine-protein kinase TEL1 | | Authors: | Xin, J. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of allosteric regulation of Tel1/ATM kinase.
Cell Res., 29, 2019
|
|
7WZW
 
 | | Cryo-EM structure of MEC1-DDC2-MMS | | Descriptor: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | Authors: | Zhang, Q, Zhang, Q. | | Deposit date: | 2022-02-19 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of Mec1/ATR kinase endogenously stimulated by different genotoxins.
Cell Discov, 8, 2022
|
|
