3IR3
 
 | | Crystal structure of human 3-hydroxyacyl-thioester dehydratase 2 (HTD2) | | Descriptor: | 3-hydroxyacyl-thioester dehydratase 2, MALONATE ION | | Authors: | Ugochukwu, E, Cocking, R, Burgess-Brown, N, Pilka, E, Muniz, J, Krojer, T, Chaikuad, A, Gileadi, O, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of human 3-hydroxyacyl-thioester dehydratase 2 (HTD2)
To be Published
|
|
3Q3V
 
 | | Crystal structure of Phosphoglycerate Kinase from Campylobacter jejuni. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Phosphoglycerate kinase, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-22 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
2F57
 
 | | Crystal Structure Of The Human P21-Activated Kinase 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, Serine/threonine-protein kinase PAK 7 | | Authors: | Eswaran, J, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Bray, J, Das, S, Savitsky, P, Smee, C, Burgess, N, Fedorov, O, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs.
Structure, 15, 2007
|
|
2BV5
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PROTEIN TYROSINE PHOSPHATASE PTPN5 AT 1.8A RESOLUTION | | Descriptor: | GLYCEROL, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE, ... | | Authors: | Debreczeni, J.E, Barr, A.J, Eswaran, J, Smee, C, Burgess, N, Gileadi, O, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-06-22 | | Release date: | 2005-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem. J., 395, 2006
|
|
2EU9
 
 | | Crystal Structure of CLK3 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Papagrigoriou, E, Rellos, P, Das, S, Ugochukwu, E, Turnbull, A, von Delft, F, Bunkoczi, G, Sobott, F, Bullock, A, Fedorov, O, Gileadi, C, Savitsky, P, Edwards, A, Aerrowsmith, C, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2005-10-28 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation.
Structure, 17, 2009
|
|
2BIJ
 
 | | Crystal structure of the human protein tyrosine phosphatase PTPN5 (STEP, striatum enriched enriched Phosphatase) | | Descriptor: | SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE, NON-RECEPTOR TYPE 5 | | Authors: | Barr, A.J, Debreczeni, J.E, Eswaran, J, Smee, C, Burgess, N, Gileadi, O, Sundstrom, M, Arrowsmith, C, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures and inhibitor identification for PTPN5, PTPRR and PTPN7: a family of human MAPK-specific protein tyrosine phosphatases.
Biochem. J., 395, 2006
|
|
7SUC
 
 | |
2GJT
 
 | | Crystal structure of the human receptor phosphatase PTPRO | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase PTPRO | | Authors: | Barr, A, Ugochukwu, E, Eswaran, J, Das, S, Niesen, F, Savitsky, P, Turnbull, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, von Delft, F, Papagrigoriou, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2C2N
 
 | | Structure of human mitochondrial malonyltransferase | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-(2-ETHOXYETHOXY)ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ... | | Authors: | Wu, X, Bunkoczi, G, Smee, C, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2005-09-29 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Different Specificities of Acyltransferases Associated with the Human Cytosolic and Mitochondrial Fatty Acid Synthases.
Chem.Biol., 16, 2009
|
|
1QZ4
 
 | |
2HE7
 
 | | FERM domain of EPB41L3 (DAL-1) | | Descriptor: | Band 4.1-like protein 3 | | Authors: | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Schiavone, L.H, Johansson, I, Hogbom, M, Karlberg, T, Kotenyova, T, Nilvebrandt, J, Norberg, P, Stenmark, P, Nordlund, P, Nilsson-ehle, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Uppenberg, J, Van den berg, S, Weigelt, J, Persson, C, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B).
J.Biol.Chem., 286, 2011
|
|
2GSE
 
 | | Crystal Structure of Human Dihydropyrimidinease-like 2 | | Descriptor: | CALCIUM ION, Dihydropyrimidinase-related protein 2 | | Authors: | Ogg, D, Stenmark, P, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Holmberg-Schiavone, L, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of human collapsin response mediator protein 2, a regulator of axonal growth.
J.Neurochem., 101, 2007
|
|
2HW4
 
 | | Crystal structure of human phosphohistidine phosphatase | | Descriptor: | 14 kDa phosphohistidine phosphatase, FORMIC ACID | | Authors: | Busam, R.D, Thorsell, A.G, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-31 | | Release date: | 2006-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First structure of a eukaryotic phosphohistidine phosphatase
J.Biol.Chem., 281, 2006
|
|
1ZDN
 
 | | Ubiquitin-conjugating enzyme E2S | | Descriptor: | SODIUM ION, Ubiquitin-conjugating enzyme E2S | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1ZKC
 
 | | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b | | Descriptor: | BETA-MERCAPTOETHANOL, Peptidyl-prolyl cis-trans isomerase like 2 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases.
PLoS Biol., 8, 2010
|
|
2A7L
 
 | | Structure of the human hypothetical ubiquitin-conjugating enzyme, LOC55284 | | Descriptor: | Hypothetical ubiquitin-conjugating enzyme LOC55284, SODIUM ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-05 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
2A4D
 
 | | Structure of the human ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
6ZFH
 
 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclopentane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclopentane]-5-one, Galactokinase, beta-D-galactopyranose | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.439 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
2ARY
 
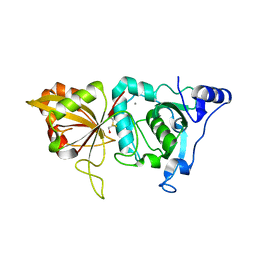 | | Catalytic domain of Human Calpain-1 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Walker, J.R, Davis, T, Lunin, V, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-22 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structures of Human Calpains 1 and 9 Imply Diverse Mechanisms of Action and Auto-inhibition
J.Mol.Biol., 366, 2007
|
|
7ABS
 
 | | Structure of human DCLRE1C/Artemis in complex with DNA - re-evaluation of 6WO0 | | Descriptor: | DNA (5'-D(*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*GP*CP*T)-3'), Protein artemis, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
3LM0
 
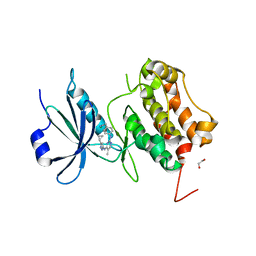 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Serine/threonine-protein kinase 17B, ... | | Authors: | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: |
|
|
7B2X
 
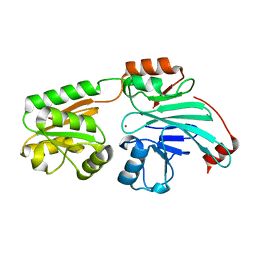 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
2C7S
 
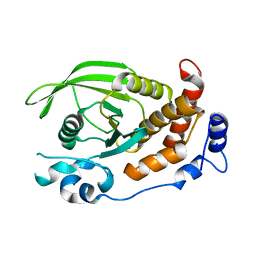 | | Crystal structure of human protein tyrosine phosphatase kappa at 1.95A resolution | | Descriptor: | ACETATE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE KAPPA | | Authors: | Debreczeni, J.E, Ugochukwu, E, Eswaran, J, Barr, A, Das, S, Burgess, N, Gileadi, O, Longman, E, von Delft, F, Knapp, S, Sundstron, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-11-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
1YRV
 
 | | Novel Ubiquitin-Conjugating Enzyme | | Descriptor: | ubiquitin-conjugating ligase MGC351130 | | Authors: | Walker, J.R, Choe, J, Avvakumov, G.V, Newman, E.M, MacKenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
6Q3X
 
 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclohexane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
