3MTS
 
 | | Chromo Domain of Human Histone-Lysine N-Methyltransferase SUV39H1 | | Descriptor: | Histone-lysine N-methyltransferase SUV39H1 | | Authors: | Lam, R, Li, Z, Wang, J, Crombet, L, Walker, J.R, Ouyang, H, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human SUV39H1 Chromodomain and Its Recognition of Histone H3K9me2/3.
Plos One, 7, 2012
|
|
6B2Q
 
 | | Dual Inhibition of the Essential Protein Kinases A and B in Mycobacterium tuberculosis | | Descriptor: | 3-methyl-1-(2-methylpropyl)butyl 4-O-beta-L-gulopyranosyl-beta-D-glucopyranoside, 5-{5-chloro-4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}thiophene-2-sulfonamide, Serine/threonine-protein kinase PknA | | Authors: | Zuccola, H.J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design To Minimize Host Kinase Interactions.
ACS Med Chem Lett, 8, 2017
|
|
6B2P
 
 | | Dual Inhibition of the Essential Protein Kinases A and B in Mycobacterium tuberculosis | | Descriptor: | 5-{5-chloro-4-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]pyrimidin-2-yl}thiophene-2-sulfonamide, Serine/threonine-protein kinase PknB | | Authors: | Zuccola, H.J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Mtb PKNA/PKNB Dual Inhibition Provides Selectivity Advantages for Inhibitor Design To Minimize Host Kinase Interactions.
ACS Med Chem Lett, 8, 2017
|
|
8E3S
 
 | |
7DY7
 
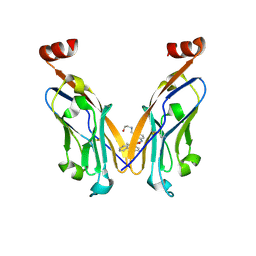 | | Discovery of Novel Small-molecule Inhibitors of PD-1/PD-L1 Axis that Promotes PD-L1 Internalization and Degradation | | Descriptor: | 2-[[3-[[5-(2-methyl-3-phenyl-phenyl)-1,3,4-oxadiazol-2-yl]amino]phenyl]methylamino]ethanol, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Wang, T.Y, Lu, M.L, Jiang, S, Xiao, Y.B. | | Deposit date: | 2021-01-20 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation.
J.Med.Chem., 65, 2022
|
|
8FZR
 
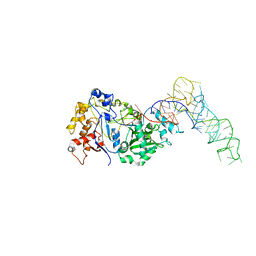 | |
6UVU
 
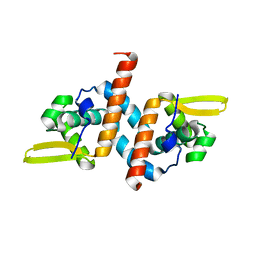 | | Crystal structure of the AntR antimony-specific transcriptional repressor | | Descriptor: | ArsR family transcriptional regulator | | Authors: | Thiruselvam, V, Banumathi, S, Palani, K, Manohar, R, Rosen, B.P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of AntR, an Sb(III) responsive transcriptional repressor.
Mol.Microbiol., 116, 2021
|
|
6THP
 
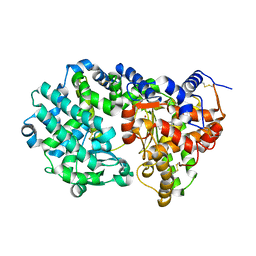 | |
8X5D
 
 | |
6KVA
 
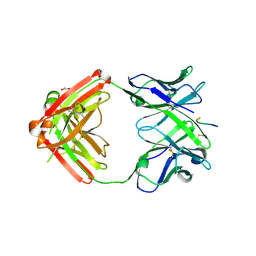 | | Structure of anti-hCXCR2 abN48-2 in complex with its CXCR2 epitope | | Descriptor: | 1,2-ETHANEDIOL, Peptide from C-X-C chemokine receptor type 2, heavy chain, ... | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
6KVF
 
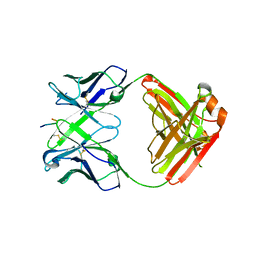 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
1BC2
 
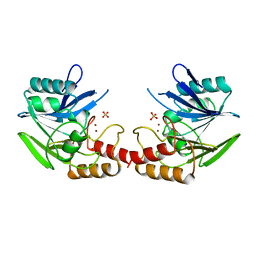 | | ZN-DEPENDENT METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE II, SULFATE ION, ZINC ION | | Authors: | Fabiane, S.M, Sutton, B.J. | | Deposit date: | 1997-04-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the zinc-dependent beta-lactamase from Bacillus cereus at 1.9 A resolution: binuclear active site with features of a mononuclear enzyme.
Biochemistry, 37, 1998
|
|
5CNA
 
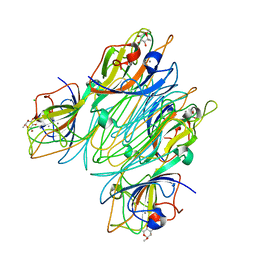 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5H0O
 
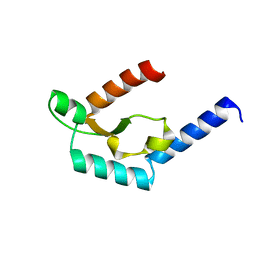 | | Crystal structure of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease with manganese ion | | Descriptor: | HNH endonuclease, MANGANESE (II) ION | | Authors: | Zhang, L.K, Xu, D.D, Huang, Y.C, Gong, Y. | | Deposit date: | 2016-10-06 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease.
Sci Rep, 7, 2017
|
|
5H0M
 
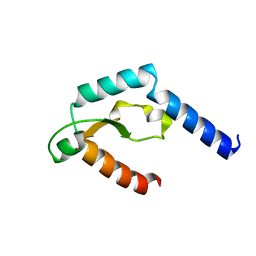 | | Crystal structure of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease with zinc ion | | Descriptor: | HNH endonuclease, ZINC ION | | Authors: | Zhang, L.K, Xu, D.D, Huang, Y.C, Gong, Y. | | Deposit date: | 2016-10-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease
Sci Rep, 7, 2017
|
|
1E07
 
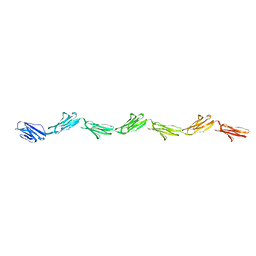 | |
1ADQ
 
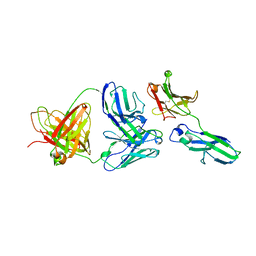 | |
2Y7Q
 
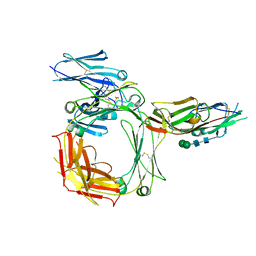 | | THE HIGH-AFFINITY COMPLEX BETWEEN IGE AND ITS RECEPTOR FC EPSILON RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR SUBUNIT ALPHA, IG EPSILON CHAIN C REGION, ... | | Authors: | Davies, A.M, Holdom, M.D, Nettleship, J.E, Beavil, A.J, Owens, R.J, Sutton, B.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Nat.Struct.Mol.Biol., 18, 2011
|
|
2WQR
 
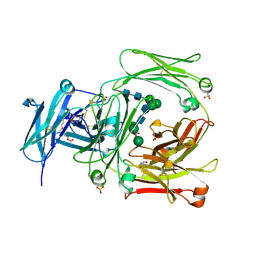 | | The high resolution crystal structure of IgE Fc | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IG EPSILON CHAIN C REGION, ... | | Authors: | Dhaliwal, B, Sutton, B.J, Beavil, A.J. | | Deposit date: | 2009-08-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Nat.Struct.Mol.Biol., 18, 2011
|
|
3BC2
 
 | |
2BC2
 
 | |
2BGA
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH7 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
2BG8
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT and 1mM TCEP-HCl were used as reducing agents. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
2BG7
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
2BG2
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in the buffer. 1mM DTT and 1mM TCEP- HCl were used as reducing agents. Cys221 is reduced. | | Descriptor: | BETA-LACTAMASE II, CHLORIDE ION, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism.
Biochemistry, 44, 2005
|
|
