2MDB
 
 | | Tachyplesin I in the presence of lipopolysaccharide | | Descriptor: | Tachyplesin-1 | | Authors: | Kushibiki, T, Kamiya, M, Aizawa, T, Kumaki, Y, Kikukawa, T, Mizuguchi, M, Demura, M, Kawabata, S.I, Kawano, K. | | Deposit date: | 2013-09-09 | | Release date: | 2014-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide.
Biochim.Biophys.Acta, 1844, 2014
|
|
2JX8
 
 | | Solution structure of hPCIF1 WW domain | | Descriptor: | Phosphorylated CTD-interacting factor 1 | | Authors: | Kouno, T, Iwamoto, Y, Hirose, Y, Aizawa, T, Demura, M, Kawano, K, Ohkuma, Y, Mizuguchi, M. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, and 15N resonance assignments of hPCIF1 WW domain
To be Published
|
|
2N92
 
 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
2E45
 
 | |
1C34
 
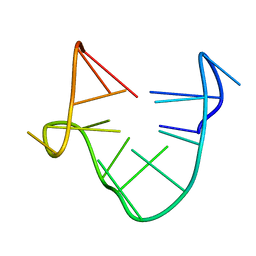 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-24 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
1C32
 
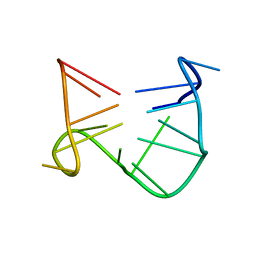 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-24 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
1C35
 
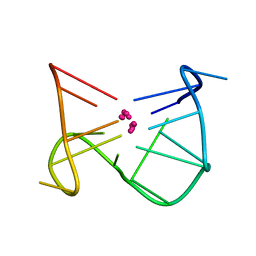 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-25 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
1C38
 
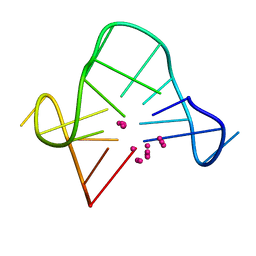 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMEDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-25 | | Release date: | 1999-08-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
2E8J
 
 | |
2DJC
 
 | | Solution structure of growth-blocking peptide of the tobacco cutworm, Spodoptera litura | | Descriptor: | growth-blocking peptide | | Authors: | Umetsu, Y, Kamiya, M, Aizawa, T, Kumaki, Y, Demura, M, Kawano, K. | | Deposit date: | 2006-03-31 | | Release date: | 2007-03-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Growth-blocking Peptide of the Tobacco Cutworm, Spodoptera litura
to be published
|
|
2DJ9
 
 | | Solution structure of growth-blocking peptide of the cabbage armyworm, Mamestra brassicae | | Descriptor: | growth-blocking peptide | | Authors: | Umetsu, Y, Kamiya, M, Aizawa, T, Kumaki, Y, Demura, M, Kawano, K. | | Deposit date: | 2006-03-31 | | Release date: | 2007-03-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Growth-blocking Peptide of the Cabbage Armyworm, Mamestra brassicae
to be published
|
|
3W01
 
 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZZ
 
 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZX
 
 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W02
 
 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W00
 
 | | Crystal structure of PcrB complexed with G1P and FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
7XB2
 
 | |
7WL3
 
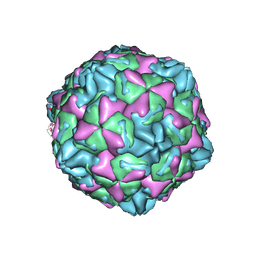 | | CVB5 expended empty particle | | Descriptor: | Capsid protein, Genome polyprotein | | Authors: | Yang, P, Wang, K. | | Deposit date: | 2022-01-12 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.
J.Virol., 96, 2022
|
|
7B54
 
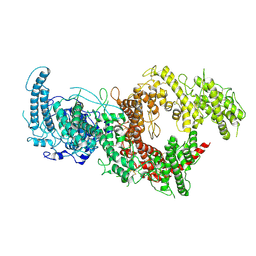 | | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | | Descriptor: | VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Dagil, R, Gourdon, P.E, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-02 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
7B52
 
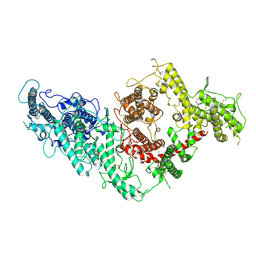 | | VAR2CSA full ectodomain | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Gourdon, P.E, Dagil, R, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
6QVD
 
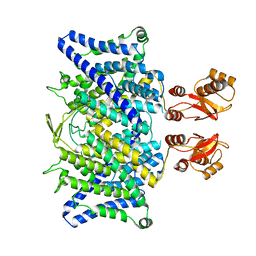 | |
6QV6
 
 | |
