4K0B
 
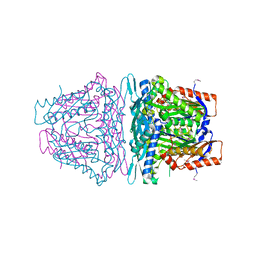 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4JJN
 
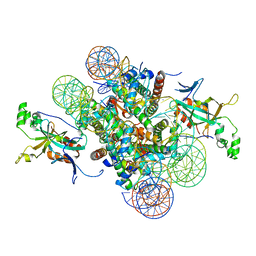 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5GWY
 
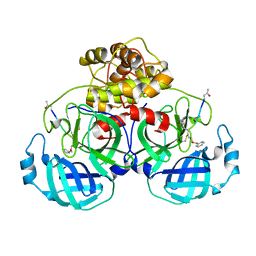 | | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, main protease | | Authors: | Wang, F, Chen, C, Tan, W, Yang, K, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structure of Main Protease from Human Coronavirus NL63: Insights for Wide Spectrum Anti-Coronavirus Drug Design.
Sci Rep, 6, 2016
|
|
2PFC
 
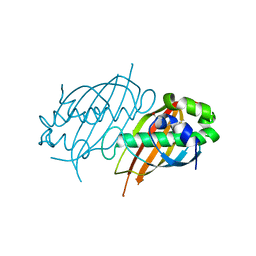 | | Structure of Mycobacterium tuberculosis Rv0098 | | Descriptor: | Hypothetical protein Rv0098/MT0107, PALMITIC ACID | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2007-04-04 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence.
Chem.Biol., 14, 2007
|
|
4J1U
 
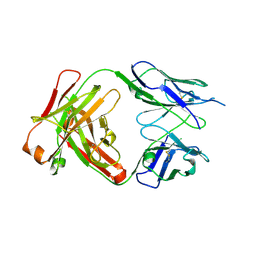 | | Crystal structure of antibody 93F3 unstable variant | | Descriptor: | antibody 93F3 Heavy chain, antibody 93F3 Light chain | | Authors: | Wang, F. | | Deposit date: | 2013-02-02 | | Release date: | 2013-03-13 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4PIW
 
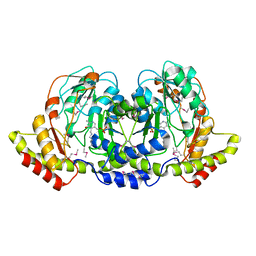 | | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12 | | Descriptor: | TDP-4-keto-6-deoxy-D-glucose transaminase family protein | | Authors: | Wang, F, Xu, W, Helmich, K.E, Singh, S, Yennamalli, R.M, Miller, M.D, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12
To Be Published
|
|
8UI7
 
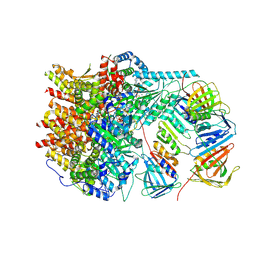 | |
8UI9
 
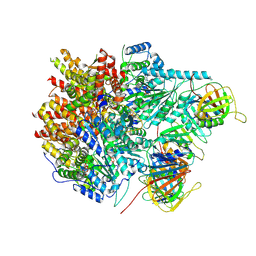 | |
8UII
 
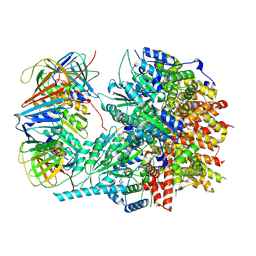 | |
8UI8
 
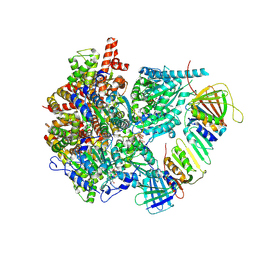 | |
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
7ESF
 
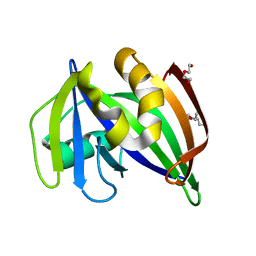 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
4FZR
 
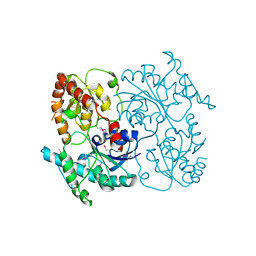 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
4G2T
 
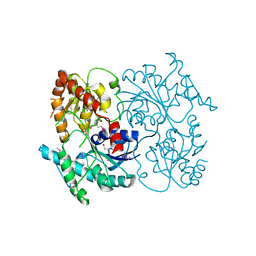 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | Descriptor: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
2HKC
 
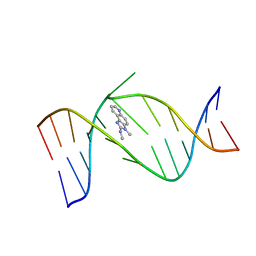 | | NMR Structure of the IQ-modified Dodecamer CTCGGC[IQ]GCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
2HKB
 
 | | NMR Structure of the B-DNA Dodecamer CTCGGCGCCATC | | Descriptor: | 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
2LQH
 
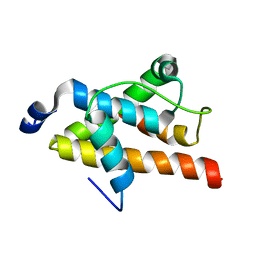 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NTJ
 
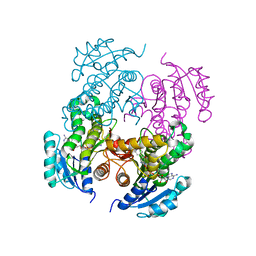 | | Mycobacterium tuberculosis InhA bound with PTH-NAD adduct | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH, {(2R,3S,4R,5R)-5-[(4S)-3-(AMINOCARBONYL)-4-(2-PROPYLISONICOTINOYL)PYRIDIN-1(4H)-YL]-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL}M ETHYL [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of thioamide drug action against tuberculosis and leprosy.
J.Exp.Med., 204, 2007
|
|
2NTV
 
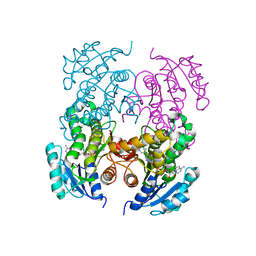 | | Mycobacterium leprae InhA bound with PTH-NAD adduct | | Descriptor: | Enoyl-[ACP] reductase, {(2R,3S,4R,5R)-5-[(4S)-3-(AMINOCARBONYL)-4-(2-PROPYLISONICOTINOYL)PYRIDIN-1(4H)-YL]-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL}M ETHYL [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Wang, F, Sacchettini, J.C. | | Deposit date: | 2006-11-08 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of thioamide drug action against tuberculosis and leprosy.
J.Exp.Med., 204, 2007
|
|
7LQH
 
 | | Cryo-EM of KFE8 thinner nanotube (class 2, 2-sub-1) | | Descriptor: | KFE8 peptide | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
7LQF
 
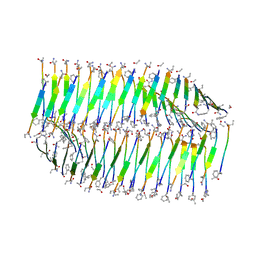 | |
7LQI
 
 | |
7LQE
 
 | |
7LQG
 
 | | Cryo-EM of the KFE8 thinner nanotube (class 1, C2) | | Descriptor: | KFE8 peptide | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Deterministic chaos in the self-assembly of beta sheet nanotubes from an amphipathic oligopeptide.
Matter, 4, 2021
|
|
