8H4I
 
 | | DHA-bound FFAR4 in complex with Gs | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4L
 
 | | DHA-bound FFAR4 in complex with Gq | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
8H4K
 
 | | GW9508-bound FFAR4 in complex with Gq | | Descriptor: | 3-(4-{[(3-phenoxyphenyl)methyl]amino}phenyl)propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Yin, H. | | Deposit date: | 2022-10-10 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of omega-3 fatty acid receptor FFAR4 activation and G protein coupling selectivity.
Cell Res., 33, 2023
|
|
6KVA
 
 | | Structure of anti-hCXCR2 abN48-2 in complex with its CXCR2 epitope | | Descriptor: | 1,2-ETHANEDIOL, Peptide from C-X-C chemokine receptor type 2, heavy chain, ... | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
6KVF
 
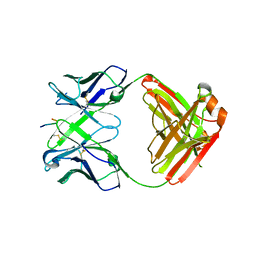 | | Structure of anti-hCXCR2 abN48 in complex with its CXCR2 epitope | | Descriptor: | Peptide from C-X-C chemokine receptor type 2, heavy chain, light chain | | Authors: | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
8FFE
 
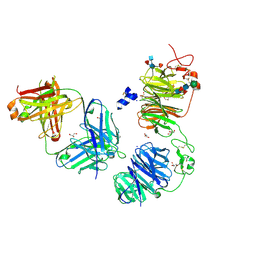 | | Crystal structure of LRP6 E1E2 domains bound to YW210.09 Fab and engineered XWnt8 peptide | | Descriptor: | GLYCEROL, Low-density lipoprotein receptor-related protein 6, SODIUM ION, ... | | Authors: | Jude, K.M, Tsutsumi, N, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5GH9
 
 | | Crystal structure of CBP Bromodomain with H3K56ac peptide | | Descriptor: | CREB-binding protein, Histone H3 | | Authors: | Xu, L. | | Deposit date: | 2016-06-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insight into CBP bromodomain-mediated recognition of acetylated histone H3K56ac
FEBS J., 2017
|
|
8X5D
 
 | |
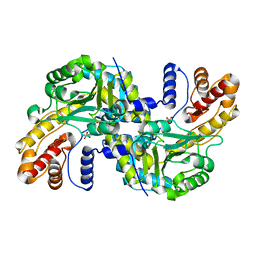
8WOU
 
 | |
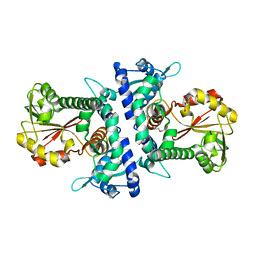
6IWT
 
 | |
4M5C
 
 | | Crystal Structure of an Truncated Transition metal Transporter | | Descriptor: | COBALT (II) ION, Cobalamin biosynthesis protein CbiM, HEXANE-1,6-DIOL | | Authors: | Yu, Y, Zhou, M.Z, Gu, J.K. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
5KF4
 
 | |
7WLC
 
 | |
7WED
 
 | |
7WEF
 
 | |
7WEA
 
 | |
7WEC
 
 | |
7WE8
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv265 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 265, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WE7
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 282, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7WEE
 
 | |
7WEB
 
 | |
7WE9
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
4M58
 
 | | Crystal Structure of an transition metal transporter | | Descriptor: | Cobalamin biosynthesis protein CbiM, NICKEL (II) ION | | Authors: | Yu, Y, Yan, C.Y, Zhang, B, Li, X.L, Gu, J.K. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
4M5B
 
 | | Crystal Structure of an Truncated Transition Metal Transporter | | Descriptor: | Cobalamin biosynthesis protein CbiM, HEXANE-1,6-DIOL, NICKEL (II) ION | | Authors: | Yu, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
7KBB
 
 | |
